* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download What are the functions of AT3G56230? AT4G18650?
Saethre–Chotzen syndrome wikipedia , lookup
Genetically modified crops wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic engineering wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene therapy wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene desert wikipedia , lookup
Epitranscriptome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
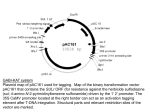
Arabidopsis thealiana AT3G56230 AT4G18650 What are the functions of them? Are they important for seed development? Spring 2004 Mariko Onozawa What is AT3G56230? FV RV ・About 1200bps. ・5’UTR. ・Three SALK lines. ・This protein containing BTB/POZ domain. What is AT4G18650? FV RV ・About 1400bps. ・5’ and 3’UTR. ・No SALK line. ・This protein is related to transcription factor and has weak similarity to TGAGG-sequence specific DNAbinding protein(TGA-2.1). Reverse Genetics mRNA Gene Phenotype mRNA *Find the pattern of mRNA concentration RT-PCR (SRB) Genechip database Gene * Find KOs of my gene Madison lines SALK lines Phenotype *Study a phenotype of plants which containing the KO Check genotypes of the plants (SALK) RT-PCR result using SRB (AT3G56230) Genechip data A border between presence and absence 3000 *RT-PCR result says that the gene which is homologous toAT3G56230 is expressed at all stages and organs of SRB. *Genechip data says that AT3G56230 is expressed only at Mature green seed, Seedling (3DAI), Roots and stem. AT4G18650 AT3G56230 2500 2000 1500 1000 control gene specific 500 St e m Ro o t s Le ave s se e dlin g (3 DAI) Po st - M at u r at io n GS M at u r e GS M id- M at u r at io n 2 4 - Hr Se e ds Ovu le s -RT Stem +RT -RT Leaves +RT -RT Axes +RT -RT Embryo +RT -RT Ovules +RT -RT Flower +RT -RT Stem +RT -RT Leaves +RT 0 Is there any KO of my gene in Madison lines? *AT3G56230 R19 FV RV I couldn’t find any KO of AT3G56230. The insertion site is upstream of the gene. (-594bp) *AT4G18650 FV I couldn’t find any KO of AT4G18650. The insertion site is downstream of the gene. R15 RV Checking phenotypes of the plants ①WT ①WT Only tubulin’s bands ①T-DNA ①T-DNA nothing Nothing ②WT, T-DNA ②WT, T-DNA Some seedlings don’t Some samples don’t have any band. have any band. Only tubilan’s bands T-DNA,homo 1,4 WT, homo 2,3,5,6 smears 1 2 3 4 5 6 WC ④WT, 1 2 3 4T-DNA 5 6 WC smears 1 2 3 4 5 6WC 1 2 3 4 5 6WC ④WT ③WT, T-DNA ③WT, T-DNA T-DNA I couldn’t find any significant difference between WT and mutant plants. T-DNA Wild Type What should be done next to study the functions? *Get more plants which have a insertion in the genes. *Observe and study the knockout plants. *Study with other knockout lines. The final goal is to answer these questions. *What is the function of the protein? *What is the structure of the protein? *How it is regulated? What we did in this class is only the beginning of the research.