* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download No Slide Title
Holliday junction wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Gene expression wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Genetic code wikipedia , lookup
Transcriptional regulation wikipedia , lookup
DNA barcoding wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Maurice Wilkins wikipedia , lookup
DNA sequencing wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Genomic library wikipedia , lookup
Restriction enzyme wikipedia , lookup
Molecular cloning wikipedia , lookup
Point mutation wikipedia , lookup
Biosynthesis wikipedia , lookup
SNP genotyping wikipedia , lookup
DNA supercoil wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Molecular evolution wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Community fingerprinting wikipedia , lookup
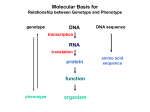
Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence Use of Restriction Fragment Length Polymorphism I. Marker locus II. Diagnostic A. Medicine B. Forensics III. Assessment of Genetic Variation A. Within and between populations B. Within and between species Restriction Mapping versus RFLP Mapping I. Restriction Mapping A. Based on physical analysis of DNA B. Based on restriction sites with no variation C. Mostly short-range (fine-scale) maps II. RFLP Mapping A. Based on recombination analysis of matings B. Based on restriction-site variation between homologous chromosomes C. Mostly longe-range (coarse-scale) maps Other Useful Approaches 1. Single Nucleotide Polymorphisms (SNPs) Individuals differ in single nucleotides (every 11 to 300 bp in interval). 2. Simple-Sequence Length Polymorphisms (SSLPs) Very short repetitive DNA sequences are more polymorphic than RFLP sequences. These are also called Variable Number Tandem Repeats (VNTRs) - Minisatellite Markers - Microsatellite Markers Simple-Sequence Length Polymorphisms 1. Minisatellite DNA These are 1 to 5 kb in length consisting of repeats 15 to 100 nucleotides in length and are identified by Southern analysis. 2. Microsatellite DNA These are tandem repeats of dinucleotides, commonly stretches of CA. 5’ C A C A C A C A C A C A C A 3’ 3’ G T G T G T G T G T G T G T 5’ These are identified by gel electrophoresis of PCR products. Molecular Basis for Relationship between Genotype and Phenotype genotype DNA DNA sequence transcription RNA translation protein function phenotype organism amino acid sequence Dideoxy DNA Sequencing Chain-terminating (dideoxy) nucleotide Use of dideoxy nucleotide in primer extension reaction will randomly arrest DNA synthesis. Dideoxy DNA Sequencing 4 different reactions are conducted, each with a different type of dideoxy nucleotide. Fragments are separated by electrophoresis. Banding pattern is used to infer the base sequence of the original template strand. Migration 3’ Sequencing Gel Using Radioactive Primer 5’ Remember… This is base sequence of synthesized strand. Reading the DNA sequence from an automatic sequencer Oligonucleotide primers can be tagged with fluorescent dyes instead of radioactive labels. A different colored dye can be used for each of the four reactions. In Search of Potential Genes Open reading frames (ORFs) are long stretches of DNA that start with ATG and end with a stop codon. A doublestranded DNA molecule has 6 possible reading frames, 3 for each strand.