* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chapter 13 Chromatin Structure and its Effects on
Messenger RNA wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Molecular cloning wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Non-coding RNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genomic imprinting wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epitranscriptome wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Ridge (biology) wikipedia , lookup
Transcription factor wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene regulatory network wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene expression wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Acetylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Community fingerprinting wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Promoter (genetics) wikipedia , lookup
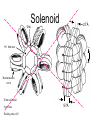
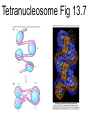
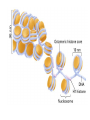
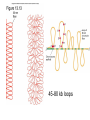
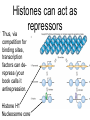
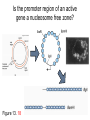
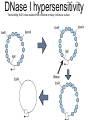
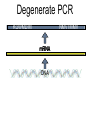
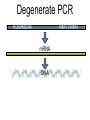
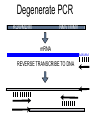
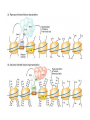
Chapter 13 Chromatin Structure and its Effects on Transcription Students must be positive that they understand standard PCR. There is a resource on the web for this purpose. Warn them before this class. Notes • Do not attempt to interpret figure 13.14. • Figure 13.18. The concentration of nucleosomes does not prevent the restriction enzyme from finding its cut sites in many molecules (in some the nucleosomes block the cut site). Histone Eukaryotes five different Histone Classes size amino acids Molecular weight H2A 150 14,000 H2B 150 13,770 H3 150 15,400 H4 100 11,340 H1 (H5) 200 21,500 Eukaryotes contain many copies of each histone gene. 10-20 in mice, 100X in Drosophila Compaction Double helix Beads On A string solenoid Snake The Solenoid PR 25 700 nm fiber Probably involve SARS Nucleosome DNA Protein Core nucleosome contains 2X H2A, 2X H2B, 2X H3 & 2X H4 Beads on a string Beads on a string packing ratio 5 Histone H1 • Outside of the core • Easiest to remove by high salt H2B extraction H2 A H3 H1 H4 H2 B DNA Solenoid 27Å DNA H1 hist one Nucleosome cor e 30 nm solenoid 6 per turn Packing ratio of 8 57Å Tetranucleosome Fig 13.7 45-80 kb loops The movie was deleted to save space. You can download it separately. Evidence that histones help to regulate gene • Xenopusexpression laevis 5s rRNA (20,000 copies) • oocyte 5S rRNA genes, expressed only in oocytes - 98% • somatic 5S rRNA genes, expressed in both oocytes and somatic cells - 2% Chromatin is required for specificity • With DNA, RNA polymerase III transcribes both well • With oocyte chromatin, both expressed • With somatic cell chromatin only somatic 5S rRNA genes expressed. Somatic cell chromatin • Inactive oocyte genes contained all 5 histones* • Active somatic genes contain only core histones • Remove H1 & oocyte genes turn on. Add it back and they turn off. pg 369 Weaver 4th ed. Nucleosome compete with transcription factors • This is too simple to allow one to modulate expression or is it? mRNA encoding genes Class II genes transcribed by RNA polymerase II • Core histones cause mild (4X) repression of gene expression • Activators do not affect this • H1 increases repression (25 to 100X) • Activators can prevent this - similar to the 5S genes. Laybourne & • Chromatin form of the Drosophila Kadonaga 1991 Krüppel gene • nuclear extract transcribes it at 25% of the maximum rate. This is 75% repression. • Interpretation 1) 100% of the genes are transcribed at a 75% rate of transcription 2) 25% of the genes are transcribed at 100% of the rate. • 25% of promoters unoccupied. How pg 370 Weaver 4th ed Histones can act as repressors • Thus, via competition for binding sites, transcription factors can derepress (your book calls it antirepression. Histone H1 Nucleosome core • • • Histone antirepression With respect to histone repression, Gal4 acts as an antirepressor. In addition, it acts as an activator. SP1 acts as both a histone antirepressor and as an activator. GAGA factor seems to only act as a histone antirepressor. De-repression or Anti-repression is the amount of transcription that you get when the histone is not interfering by hiding the promoter. • • • Histone antirepression With respect to histone repression, Gal4 acts as an antirepressor. In addition, it acts as an activator. SP1 acts as both a histone antirepressor and as an activator. GAGA factor seems to only act as a histone antirepressor. Yaniv saw that some transcriptionally active SV40 virus DNAs had nucleosome free zones. Fig. 13.21 Weaver 3rd ed. Figure 13.17 Is the promoter region of an active gene a nucleosome free zone? Figure 13. 18 BamHI BamHI BamHI BglII BglII BglII DNase I hypersensitivity Transcribing SV40 virus isolated from infected monkey cell tissue culture. Two fragments Figure 13. 20 Gel electrophoresi s & Southern Blotting This is a Southern Blot. DNase I hypersensitivity in Chromatin lectures Is this caused by the promoter or something else near the promoter? Can this occur with the promoter at a different location? Try a modified SV40 that has a second promoter inserted. Does transcription cause this or is it caused by something else that binds the promoter? Do we need to have transcription for this to occur? Make a nuclear lysate that supports transcription & chromatin assembly. Then deplete it for RNA polymerase II. Or starve for nucleotides. Is this peculiar to the SV40 promoter. Try a plasmid with a completely different type of promoter. Does the promoter have to be active for this to happen? Try this with a promoter that can be turned on and off. eg. Tet-on. Why does the smaller band disappear? *Less intense because amount of probe that it can collect is smaller. Test w/2 probes of the same size. *Less abundant, because transcription is going that way and polymerarse might expand the nucleosome clear zone to the left. Test by reversing the direction of the plasmid. What is the origin of the 100% band. Does it confound our interpretation? Histone acetylation • amino groups of lysine side chains • unacetylated histones tend to repress transcription • acetylated histones tend to activate transcription • Histone acetyl transferase (HAT) • Histone deacetylase - see Figure 13.23 for how to detect them. Histone acetylation • First discovered by Vincent Allfrey in 1964 takes ‘till 1996 (Brownell & Allis) to purify a HAT • Tetrahymena has heavily acetylated histones. • Take macronuclei extracts. SDS gel electrophoresis in gel impregnated with histones. Soak in radiolabeled acetylCoA. Wash. Fluorography. You don’t have this slide. How to purify a HAT • What do you know about it? Acetylates histones • What does this knowledge provide you? A way to assay for its presence • What is the advantage of using a gel? Demonstrates that one is detecting the presence of a single enzyme. Tells you the relative size of the enzyme. heated chemical inactivation BSA no histones no protein Now purify it. • Standard biochemical techniques to purify p55. • Can use the assay to tell where it is. • Once pure get partial amino acid sequence. Very small number of amino acids. Degenerate PCR KGWMDIM NMVTMMV mRNA DNA Degenerate PCR KGWMDIM NMVTMMV mRNA DNA Degenerate PCR KGWMDIM NMVTMMV mRNA AAAAAAA REVERSE TRANSCRIBE TO DNA Degenerate PCR KGWMDIM NMVTMMV Codon Usage by amino acid. F L S Y TTT TTA TCT TAT TTC TTG TCC TAC CTT TCA CTC TCG CTA AGT CTG AGC I ATT ATC ATA M ATG T ACT ACC ACA ACG N AAT AAC C TGT TGC W TGG P CCT CCC CCA CCG H CAT CAC Q CAA CAG R CGT CGC CGA CGG AGA AGG K AAA AAG V GTT GTC GTA GTG A GCT GCC GCA GCG D GAT GAC E GAA GAG G GGT GGC GGA GGG Degenerate PCR KGWMDIM NMVTMMV UPPER STRAND Primer cocktail K G W M 5' AA(AG) GG(N) TGG ATG 2 X 4 X 1 X 1 X D I GA(TC) 2 X M AT(TCA) ATG 3' 3 X A 21 mer primer with 48 fold degeneracy. 1 = 48 Degenerate PCR KGWMDIM NMVTMMV LOWER STRAND Primer cocktail N M V T M M V 5' AA(TC) ATG GT(N) AC(N) ATG ATG GT(N) 3' Take the reverse complement for the lower strand. The reverse complement is merely the opposite strand in a DNA helix also written in the 5' to 3 direction. 5' (N)AC 4 X CAT 1 X CAT 1 X (N)GT (N)AC 4 X 4 X CAT 1 X (AG)TT 3' 2 = 128 Degenerate PCR LOWER STRAND Primer cocktail N M V T M M V 5' AA(TC) ATG GT(N) AC(N) ATG ATG GT(N) 3' Take the reverse complement for the lower strand. The reverse complement is merely the opposite strand in a DNA helix also written in the 5' to 3 direction. 5' (N)AC CAT 4 X CAT 1 X 1 X (N)GT (N)AC 4 X 4 X CAT (AG)TT 3' 1 X 2 = 128 If we wish to have a less degenerate cocktail what can we do? Let's shave off the 5' end by one base to reduce the degeneracy to 32 fold. 5' AC CAT CAT Degeneracy is 1 X 1 X 1 X (N)GT 4 X (N)AC 4 X CAT 1 X (AG)TT 3' 2 = 32 fold degeneracy. Degenerate PCR D N F N R Q K Q K L G G E D L F M T E E Q K K Y Y N A M K K L G S K K GAYAAYTTYAAYMGNCARAARCARAARYTNGGNGGNGARGAYYTNTTYATGACNGARGARCARAARAARTAYTAYAAYGCNATGAARAARYTNGGNWSNAARAARG 2 2 2 2 6 2 2 2 2 6 4 4 2 2 6 2 1 4 2 2 2 2 2 2 2 2 4 1 2 2 6 4 6 2 2 L G G E D L F YTNGGNGGNGARGAYYTNTTY 6 4 4 2 2 6 2 = 4608 D N F N R Q K ...........................................K K Y Y N A M GAYAAYTTYAAYMGNCARAAR..........................................AARAARTAYTAYAAYGCNATG 2 2 2 2 6 2 2 = 384.....................................2 2 2 2 2 4 1 = 128 Ambiguous Bases A-adenine B-not A C-cytosine G-guanine H-not G K-G or T M-A or C N-A, C, G or T R-A or G S-C or G T-thymine U-uracil=T V-not T W-A or T Y-C or T - = gap Screen a cDNA library • DNA copies of every mRNA made by a cell are produced and cloned into a bacterial vector. • Screening. • Google. 5’-RACE Figure 4.16 HAT type A • Have bromodomain • Binds aceylated lysines. So HAT A’s can recoginze partially acetylated histone tails. • Examples p55, Gcn5p CBP/p300, TAF250 Acetylation continued • Acetylation of histone tails neutralizes some of the positive charge, causing them to relax their grip on the DNA. • Reduces nucleosome cross-linking. That is; the interaction between histones in neighboring nucleosome. eg. basic n-terminal tail of H4 in one nucleosome and an acidic pocket in H2A-H2B dimer in the next nucleosome Acetylation continued • Also some TFs recognize acetylated histones. eg. TAFII250 has a double bromodomain and recognizes low level acetylated histones. Once bound it is a HAT and increases acetylation. • low level acetylation of histones occurs in inactive chromatin. END Take home • The presence of nucleosomes can interfere with the binding of TFs to enhancers and with the preinitiation complex to the promoter. = Repression • When other proteins (simple TFs) are bound to the DNA they can prevent the histones from binding. This is competitive inhibition of histone binding. Called derepression or antirepression in your book.