* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Effect of Aminoguanidine (Inducible Nitric Oxide Synthase Inhibitor
Proteolysis wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Biochemical cascade wikipedia , lookup
Paracrine signalling wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Signal transduction wikipedia , lookup
Biosynthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Epitranscriptome wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Lipid signaling wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Metalloprotein wikipedia , lookup
Gene expression wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Expression vector wikipedia , lookup
Biochemistry wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
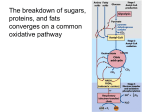
Pyruvate Dehydrogenase Kinases ABSTRACT Pyruvate dehydrogenase complex (PDC) is composed of copies of 4 different subunits (α2β2) including 3 catalytic enzymes; pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and dihydrolipoamide dehydrogenase (E3), two regulatory enzymes; pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase and E3 binding protein. PDK (PDC inhibitor) consists of 2 subunits. PDK2, PDK3 and PDK4 share same molecular weight 45KDa, while only PDK1 has molecular weight of 48KDa. PDK1 has 436 residues. PDK2 and PDK4 have 407 residues and PDK3 has 406 residues. PDK isomers contain poorly conserved N-terminal domain and highly conserved C-terminal domain. PDK4 levels increase in response to starvation, insulin resistance and diabetes. Glucocorticoids, forkhead box other factors, peroxisome proliferator-activated receptors and estrogen-related receptors induce PDK gene expression. Insulin suppresses PDK4 gene expression. PDKs inhibitors used to treat cancer, diabetes, lactic acidosis and cardiac diseases. Keywords: Pyruvate dehydrogenase complex, Pyruvate dehydrogenase kinases, Pyruvate dehydrogenase kinases inhibitors Introduction The pyruvate dehydrogenase complex (PDC) catalyzes the irreversible oxidative decarboxylation of pyruvic acid into acetyl CoA. The control of PDC appears to play a central role in shifting the carbohydrate metabolism to fat metabolism. Within mammals no pathways exist for the synthesis of pyruvate from acetyl CoA, therefore the control of the PDC reaction is crucial to avoid depletion of carbohydrate derived carbon skeletons which cannot be resynthesized by the body [1]. This reaction links the utilization of glycogen, glucose and lactate with the citric acid cycle as well as fatty acid oxidation and synthesis, since acetyl CoA can be used for fatty acid and cholesterol synthesis in the liver and fatty acid synthesis in adipose tissue. Prolonged fasting or high fat diets inactivate the PDC in order to minimize loss of pyruvate carbon to conserve it for maintaining the blood glucose levels needed through gluconeogensis. Another important reason for inactivating the PDC is to promote the usage of fatty acids as their primary energy source in many tissues as possible. This in turn conserves body protein because amino acids are not oxidized for energy and less amino acid carbon has 1 to be used for glucose synthesis [2,3]. All these observations indicate that, regulating PDC is an important step in fuel selection for energy production in animals during different nutritional and hormonal states. Thus, the regulation of PDC is critical in glucose and fatty acids metabolism. Pyruvate dehydrogenase complex (PDC): structure and function PDC is located on the mitochondrial inner membrane, where it oxidatively decarboxylates pyruvate to acetyl CoA and CO2 coupled with the reduction of NAD+ to NADH+H+ [4,5]. The PDC multienzyme complex (9.5 megadalton) composed of multiple copies of 4 different subunits (α2β2) including 3 catalytic enzymes; pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and dihydrolipoamide dehydrogenase (E3), two regulatory enzymes; pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatise (PDP) and E3 binding protein (E3BP) [6]. The role and integration of these subunits is poorly understood but the α-subunit is known to possess the serine residues that are the target for covalent modification by the intrinsic regulatory kinase and phosphatase system, responsible for regulation of PDC activity [7]. Dihydrolipoamide dehydrogenase (E3) is a FAD-containing flavoprotein which contains the single disulphide bridge that undergoes oxidation and reduction during catalysis. Dihydrolipoamide acetyltransferase (E2) forms the structural hub to which the E1, E3 and the intrinsic regulatory phosphatase and kinase bind [1]. PDC enzymes decarboxylate and dehydrogenate pyruvate in presence of thiamine pyrophosphate (TPP), free CoASH and NAD+ cofactors [8]. TPP plays the role of positive allosteric effector of pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase multienzyme complexes. It causes conformational changes of the complexes and increases their affinity to substrates [9]. E1 is made up of E1α and E1β subunits. E1α has pyruvate dehydrogenase activity and is responsible for decarboxylation of pyruvate. Each PDC is composed of 216 subunits; 60 E2, 60 E1α, 60 E1β, 12 E3BP and 24 E3 [8]. E2 subunit is responsible for transfer of acetyl group to CoA while E3 subunit transfers electrons to NAD+ forming NADH+H+ [6]. E2 forms the core of the complex, while E1 is the enzyme responsible for decarboxylation of pyruvate. E1 binds noncovalently to an E1-binding domain (E1BD) on E2 as heterotetrameric α2β2 structure. E3 is the enzyme responsible for oxidation of reduced lipoyl groups of E2 and subsequent production of NADH+H+ and it binds to the complex by E3BP, as twelve E3- 2 binding proteins anchor E3 homodimers to each PDC [8]. Each complex consists of a core of 60 E2 subunits, to which 30 E1 subunits are attached (at the E1-binding domain of E2). Six to twelve E3 subunits are tightly bound to E2 core through E3-binding protein (Figure 1). E1 component is responsible for catalysing the rate-limiting step of the overall PDC reaction [7]. E1 catalyses decarboxylation of pyruvate and the subsequent reductive acetylation of lipoyl moiety of E2. This enzyme then catalyses the transfer of acetyl group to the free CoASH cofactor [1]. Reduced lipoyl moiety of E2 is then reoxidised by E3, with NAD+ acting as the final acceptor of the reduced electron (Figure 2) [10]. Although PDC activity is directly inhibited by the acetyl CoA and NADH+H+, the two by-products of fatty acid and pyruvate oxidation, covalent modification of the PDC at the serine residues appears to be more important for regulating the PDC activity [8]. Two regulatory enzymes are involved in this covalent modification, the PDK which inactivates the PDC through phosphorylation and the PDP which activate the PDC through dephosphorylation. Four PDK isoenzymes (PDK1, PDK2, PDK3 and PDK4) and two PDP isoenzymes (PDP1 and PDP2) are expressed in mammals. The phosphorylation and the dephosphorylation occur at 3 serine residues number 203, 264 and 271 of the α-subunit of the E1 element of the PDC [8,11]. Regulation of pyruvate dehydrogense complex PDC is regulated via both short-term and long-term mechanisms. The regulation of PDC within a few minutes to hours is termed short-term regulation, while regulation that occurs at transcriptional level within days to weeks is long-term [12]. With respect to maintenance of glucose homeostasis especially during transition between fed and fasted states, PDC regulation via short-term mechanism is highly critical. The short-term regulation of PDK involves direct modification of its activity by the substrates (pyruvate, NAD+ and CoA) and the products (NADH+H+ and acetyl CoA) of the PDC reaction (Figure 3). Pyruvate inhibits PDK activity by binding to an allosteric site on PDK [13,14]. Furthermore, the effect of pyruvate to PDK is synergistic with the effect of ADP [13]. It is believed that the binding of pyruvate to the PDK-ADP complex results in conformational changes which prevent release of the ADP molecule from the active site of PDK. This ensures that PDK remains in an inactive form [14,15]. NADH+H+ and acetyl CoA 3 induce PDK activity (Bowker-Kinley et al., 1998) by binding to inner lipoyl domain (L2) of PDC [16]. The E1 subunit of PDC is inactivated through phosphorylation by PDK and reactivated via dephosphorylation by PDP [17-19]. PDK phosphorylates the E1 subunit of PDC on serine residues that are located on 3 different sites. These are designated site 1 (Ser 264), site 2 (Ser 271) and site 3 (Ser 203). Half of the site reactivity during phosphorylation of all 3 sites has been reported, which implies that a maximum of only 3 of the 6 potential phosphorylation sites (3 potential sites for each E1 subunit) can be phosphorylated [20]. Analysis of relative initial rates of phosphorylation of each site on E1 by use of rat heart complex demonstrated that site 1 phosphorylation is the most rapid and site 3 phosphorylation the least rapid [21]. Construction of mutant human E1s with single functional phosphorylation sites has revealed that, phosphorylation of each site alone can cause PDC inactivation [20]. However, to reiterate the results obtained with purified mammalian PDC[21], phosphorylation of site 1 is 4.6 fold more rapid than that of site 2 and 16 fold more rapid than that of site 3 [22]. Studies using recombinant proteins with mutations in the 3 phosphorylation sites have also revealed that dephosphorylation of sites 1, 2 and 3 occurs randomly [22]. This implies that, in functional terms, phosphorylation of sites 2 and 3 could retard the rate of dephosphorylation of all 3 sites through site competition for PDP. Multisite phosphorylation of E1 leading to impaired reactivation by PDP has been proposed as one possible explanation for the delay in PDC reactivation that is found in liver, heart and oxidative skeletal muscle on re-feeding after starvation [20,23]. The E1α mutants were constructed by converting the serine residues at different phosphorylation sites to alanine, leaving only one functional phosphorylation site at a time [22]. They reported that phosphorylation of each site alone results in the complete inactivation of PDC [18,22,24]. These sites are believed to define PDK species specificity through a phosphorylation-dephosphorylation cycle (Figure 3). The long-term regulation of PDK (Figure 4) occurs at the gene expression level and is believed to be a part of an adaptive response to conditions of decreased glucose utilization and increased fatty acid oxidation [25]. This state is induced by nutrition and hormonal changes 4 [26]. For example, starvation [26-28], high fat feeding [29] and diabetes [27,29,30] lead to high PDK4 gene expression. Active regulation of PDC by phosphorylation Not surprisingly given its central role in metabolism, PDC is under tight and complex regulation, which includes regulation by reversible phosphorylation in response to the availability of glucose. In humans, PDC activity is inhibited by site-specific phosphorylation at the 3 serine sites on the E1α subunit, which is catalyzed by four different pyruvate dehydrogenase kinases (PDK1-4). Each of the four kinases has a different reactivity for these 3 sites. Interestingly, phosphorylation at any one site leads to the inhibition of the complex in vitro. Two pyruvate dehydrogenase phosphatases (PDP1 and PDP2) dephosphorylate the E1α and activate the enzyme. Each of PDK and PDP is under transcriptional control in response to different cellular stress events (Figure 5) [3]. As an example of the transcriptional regulation, expression of PDK4 is suppressed under basal conditions in most tissues by maintaining relevant histones in deacetylated state but is induced, but its expression is increased during starvation by glucocorticoids that reacetylate these histones, particularly in heart, skeletal and other muscle tissues, kidney and liver. PDK4 is also up-regulated by a high fat diet and extended exercise. Insulin inhibits PDK4 expression. The levels of PDK4 are also regulated to peroxisome proliferator activator receptor (PPAR) transcription factors. Importantly, in diabetes caused by either insulin deficiency or insulin insensitivity, the uninhibited PDK4 overexpression prevents glucose oxidation. In contrast, the levels of PDK1 are sensitive to O2 levels and under regulation by the transcription factor hypoxia-inducible factor- (HIF-1α). An increase in the level of PDK1 is a key part of the so-called Warburg effect, a switch from oxidative to glycolytic ATP production that characterizes cancer cells [31]. PDK isomers Table 1 shows general data about PDK isoenzymes (EC number 2.7.11.2) as retrieved from the genebank databases. To date, 4 isomers of PDK have been identified and cloned. These are PDK1 [32], PDK2 [33], PDK3 [34] and PDK4 [35]. PDK isoenzymes exhibit tissue-specific expression patterns [16]. PDK1 expression has been detected in heart [26,36], pancreatic islets [37] and skeletal muscle [38]. PDK2 is expressed ubiquitously with 5 particularly high expression in heart, liver and kidney [16], while PDK3 is expressed in testes, kidney and brain [39]. PDK4 is highly expressed in several tissues, including heart [26,40], skeletal muscle [29,38], liver [41], kidney [16,37] and the pancreatic islets [37]. The distribution of the four PDK isoforms between tissues suggests that there exists a need for a differing activation of PDC between these tissues. PDK2 mRNA has been found to be highly expressed in most human tissues [33]. It is the most sensitive isoenzyme to ATP and also the most sensitive to inhibition by dichloroacetate (DCA) [16]. It has been suggested, by the same group, that PDK2 is the major PDK isoform involved in control of mammalian PDC activity, due to its profuse expression within most tissues. PDK3 possesses the highest specific activity of all PDK isoforms [16]. It has been suggested to have a specific role within the testes, although that has not yet been clarified. There is 66-74% homology in the primary structures between the four PDK isoenzymes. PDK3 and PDK4 are the most distinct and PDK1 and PDK2 the most conserved [33]. Each PDK isoform is extremely similar in sequence between species and PDK1 and PDK2 are >95% identical between rat and human [11]. The PDK2, PDK3 and PDK4 share the same molecular weight which corresponds to a 45 KDa, while only PDK1 has molecular weight of 48 KDa [11]. Classification and structure of PDK PDKs, together with branched-chain α-ketoacid dehydrogenase kinases (BCKDKs), form a novel family of eukaryotic protein kinases, namely the mitochondrial protein kinases (mPKs) [32,42]. These are serine-specific protein kinases which share more similarity with prokaryotic protein kinases, despite being eukaryotic. PDKs belong to the ATPase/kinase superfamily which consists of bacterial histidine kinases (EnzV and CheA) and ATPases (hsp90 and DNA gyrase B) [43], whereas eukaryotic protein kinases have a different molecular signature [32,44]. PDK consists of 2 subunits, the catalytically active α-subunit and the regulatory βsubunit. Four isoenzymes of PDKs (PDK1, PDK2, PDK3 and PDK4) have been identified in mammalian tissues [16,35,45]. The catalytic subunit of eukaryotic serine/threonine kinases contains 8 subdomains which play an essential role in the functioning of the enzyme through involvement in the binding and orientation of ATP. They also partake in the transfer of the γphosphate from ATP to the acceptor hydroxyl residue (Ser, Thr or Tyr) of the protein substrate [44]. 6 The primary structure of PDK was determined for the first time by isolating a cDNA encoding a 48 KDa form of PDK, later termed PDK from a rat heart cDNA library [32]. Based on analysis of the deduced amino acid sequences and crystal structures studies, it had been suggested that PDK does not belong to the eukaryotic serine/threonine protein kinases but rather it resembles the prokaryotic histidine protein kinases. Therefore, it had been assigned to the ATPase/kinase superfamily (composed of bacterial histidine protein kinases, DNA gyrases and molecular chaperone Hsp90) [15, 16,32]. Primary sequence analysis of PDK isomers shows that they contain poorly conserved N-terminal domain and highly conserved C-terminal domain. The C-terminal domain contains 3 subdomains referred to as N-box (Glu-x-x-Lys-Asn-x-x-Ala), G1-box (Asp-x-Gly-x-Gly) and G2-box (Gly-x-Gly-xGly) [15,32]. These subdomains are present throughout ATPase/kinase superfamily. Furthermore, PDK isomers show high sequence conservation among different eukaryotic species, for example rat and human PDK2 proteins share greater than 95% identity in their primary amino acids sequence. The total number of amino acid residues varies among different PDK isomers, for example PDK1 has 436 residues, PDK2 and PDK4 have 407 residues and PDK3 has 406 residues [35]. The G-boxes contain glycine rich loops which are characteristic of nucleotide binding domains [46]. Their nucleotide binding role was confirmed by resolving the structure of rat PDK2 that was crystallised with a bound molecule of ADP [15]. The residues in G1-box served as the basis of adenine specific interaction between ADP and the kinase. The G1-box is part of a loop connecting α helices 10 and 11 and it forms one side of the binding pocket with the G2-box forming the other side [15]. The G-boxes together with the N-box serves as a unique ATP binding fold. The amino acids in N-box play an important role in PDK activity [47,48] and an ATP lid [14,49]. The N-terminal domain is made up of 8 α-helices connected by flexible loops and this domain begins at residue 1 and ends at residue 179. The N-terminal domain contains conserved subdomain called an H-box (x-Asn-x-His-x-Asp) (Figure 8) which was initially predicted to be the site of PDK autophosphorylation [43]. The α-helices form a 4 helix bundle that resembles histidine phosphoryl-transfer (HPt) domain of the twocomponent systems. In a two-component signal transduction pathway, a histidine kinase catalyses the first step where sensor domain responds to a signal by autophosphorylation of a histidine residue within transmitter domain. This is then relayed by transferring phosphate 7 group to aspartic acid residue within the receiver domain of a cognate response regulator protein which is usually a transcription factor. However, in PDKs, the H-Box in N-terminal domain was proved not to be the site of PDK autophosphorylation, since autophosphorylation was mapped to serine residues in this domain [50]. Similarly, the apparent molecular masses of the PDKs determined by polyacrylamide gel electrophoresis vary such that mature PDK1 corresponds to a 45 KDa subunit, whereas mature PDK2, PDK3 and PDK4 correspond to a 48 KDa subunit. In the PDK family, PDK2 [15] and PDK3 [14] are the only isomers with crystal structures that have been solved. PDKs are reported to be homodimers with each monomer folding into a well defined 45 KDa subunit [14,15]. Figure 6 shows the ribbon representation of the structure of PDK isoenzymes as retrieved from the National Center of Bioinformatics-USA (NCBI). Reaction mechanism of PDK All studied and characterized histidine kinases transfer phosphate from an autophosphorylated histidine residue to an aspartate residue resulting in transduction of cellular signals. PDK contains two prototypical component histidine motifs [32] and His115 was presumed to be a site of autophosphorylation and a phosphor-His catalytic intermediate in PDKs. Treatment of PDK by histidine modifying reagents diethylpyrocarbonate and dichloro(2,2':6',2'-terpyridine)-platinum (II) dihydrate completely abrogates PDK activity [51]. This supported an essential role of His115 in PDK catalytic activity. This theory was later disproved by solving the crystal structure of PDK2 [15]. The determination of the structure of PDK2 suggested that His115 is solvent inaccessible making it unlikely to partake in a phosphorhistidine intermediate. Furthermore, His115 plays an important role in making a number of crucial structural hydrogen bonds [15]. Currently, the site of autophosphorylation of PDK is not clearly defined. The mutation of Leu 234 and Glu 238 amino acid residues to alanine resulted in a complete loss of PDK activity [52,53]. The asparagine and lysine residues which are also located in the N-box were found to interact directly with ATP [15]. According to the PDK2 structure, the lysine residue in the N-box is oriented towards the ATP binding site. It was therefore hypothesised that this lysine residue is involved in the catalysis of PDK. This hypothesis was strengthened by the PDK mutated at this lysine residue [53]. The conserved amino acids of the N-box (Glu and Lys) participate in binding of Mg2+ via a water molecule 8 that coordinates the Mg2+ with asparagine completing the magnesium octahedral sphere [53]. The proposed role of Glu 238 was strengthened by observing that, in the other members of the ATPase/kinase superfamily such as topoisomerase, the amino acids that correspond to Glu 238 participate indirectly in the binding of Mg2+ via a water molecule that coordinates the Mg2+ [54,55]. The base catalyst is proposed to be either located in the ATP lid or in the substrate. Kinetics of PDKs PDKs phosphorylation sites PDK isoenzymes show different kinetic parameters for phosphorylation site specificity in the regulation of PDC activity. The phosphorylation of α-subunit of the E1 element of the PDC, occurs at three specific serine residues, serine-264 (designated phosphorylation site 1), serine-271 (designated phosphorylation site 2) and serine-203 (designated phosphorylation site 3) [8,11,23]. PDC can be inactivated by phosphorylation of each site alone, although phosphorylation of site 1 is most rapid and site 3 phosphorylation least rapid. All the four PDKs isoforms exhibit higher activity towards site 1 of free E1 compared with sites 2 and 3. PDK2 exhibits the highest activity and PDK3 the lowest activity towards site 1. Hence, PDK2 activity may account for much of the short-term inhibition of PDC observed on the transition from the fed to the fasted state through phosphorylation of site 1. PDK4 exhibits a much higher activity towards site 2 compared with PDK1, PDK2 and PDK3 [56]. All four PDKs can phosphorylate sites 1 and 2 but site 3 can be phosphorylated only by PDK1, this was demonstrated using recombinant mutant proteins of the E1 element of the PDC with a single functional phosphorylation site [28,56]. All four PDK isomers have been shown to be capable of phosphorylating PDC in vitro. However, the relative catalytic activity of the PDK isozymes towards the E1 subunit of PDC varies, with PDK2 having the highest activity, followed by PDK4, PDK1 and PDK3 [16]. Further, the relative specificities of the PDK isoforms towards the three phosphorylation sites of the E1PDC subunit differ [56]. PDK isoforms are capable of phosphorylating sites 1 and 2 and only PDK1 is able to phosphorylate site 3 (Figure 3). Site 1 has been proposed to be the primary site for inactivation of PDC since site 1 phosphorylation is the most rapid and it is specific to PDK1 [56]. PDK4 showed highest activity towards site 2 when compared to the other isomers with PDK1, PDK2 and PDK3 [28,56]. The observation that the activation of 9 PDC by PDP is much slower when site 2 is phosphorylated [23] led to the hypothesis that, the phosphorylation of site 2 is primarily for slowing down the rate of re-activation of PDC by PDP [56]. PDC activity is also regulated through feedback loop. For example, the products of PDC reaction (e.g. NADH+H+ and acetyl CoA) inactivate PDC by increasing PDK activity [23,57]. The completely dephoshporylated PDC showed a positive cooperativity of pyruvate binding sites at low substrate concentrations, yet partial phosphorylation of the complex (to 35% reduction of its activity) abolished the cooperativity [58]. Specificity of PDKs Isoform-specific differences in kinetic parameters, regulation and phosphorylation site specificity introduce variations in the regulation of PDC activity in differing endocrine and metabolic states. Although all the four PDKs isoenzymes can phosphorylate and inactivate PDC in vitro, the relative catalytic activity of recombinant isoenzymes toward wild-type E1 varies (PDK2 < PDK4 < PDK1 < PDK3) [16]. In addition, individual recombinant PDK isoenzymes differ in their acute regulation by metabolites [16]. Recombinant PDK2 is the most sensitive (Ki=0.2mM) to inhibition by the pyruvate analog dichloroacetate (DCA) [16] (Figure 7). In contrast, PDK4 is relatively insensitive to suppression by DCA [16] but more responsive to an increased NADH+H+ to NAD+ concentration ratio than PDK2 [16] (Figure 7). However, further addition of acetyl CoA activates PDK2 but PDK4 does not show activation above that seen with NADH+H+ alone [16] (Figure 7). The specificity of the four mammalian PDKs toward the three phosphorylation sites of E1 has been investigated using recombinant E1 mutant proteins with only one functional phosphorylation site [28,56]. All four PDKs phosphorylate sites 1 and 2, however, site 3 is phosphorylated only by PDK1 [28,56] (Figure 7). Although PDK is activated by binding to E2 [59], all four PDKs can phosphorylate sites 1 and 2 and PDK1 can phosphorylate site 3 in the absence of E2-E3 binding protein [56]. All four PDKs exhibit higher activity toward site 1 of free E1 compared with the other two phosphorylation sites, PDK2 exhibiting the highest activity and PDK3 the lowest activity toward site 1 [56]. In contrast, in the free form, PDK4 exhibits a much higher activity toward site 2 compared with PDK1, PDK2 and PDK3 [56]. Functional significance of PDK isoform shifts 10 PDK4 is a lipid status-responsive PDK isoform, facilitating faty acids oxidation by sparing pyruvate for oxaloacetate formation [3]. In heart and skeletal muscle, increased entry of pyruvate into the tricarboxylic acid cycle (TCA) cycle as oxaloacetate facilitates entry of acetyl CoA derived from fatty acids -oxidation into the TCA cycle through increased citrate formation. In turn, citrate formation acts as a signal of fatty acids abundance to suppress glucose uptake and glycolysis. Furthermore, by maintaining acyl CoA removal by oxidation, upregulation of PDK4 would be predicted to allow continued uptake oxidation, preventing long chain fatty acyl CoA accumulation in the cytoplasm, where it would be predicted to exert deleterious effects on function. Skeletal muscle enhanced PDK4 protein expression after prolonged starvation is associated with a rightward shift in the sensitivity curve for suppression of PDK activity by pyruvate and attenuation of the maximal response of PDK to suppression by pyruvate [36]. A similar shift in sensitivity of PDK activity to suppression by pyruvate is observed in response to prolonged starvation in the heart [60]. Thus the relative insensitivity of PDK to suppression by pyruvate in heart and skeletal muscle evoked by prolonged starvation [36,60] correlates with selective increases in PDK4 protein expression [26,61]. Insulin resistance induced by prolonged high fat feeding also leads to a reduction in the sensitivity of skeletal muscle PDK to inhibition by pyruvate [29] Thus it appears that PDK isoform switching toward increased PDK4 expression can be associated with altered regulatory characteristics of PDK in vivo, as would be predicted from studies with the recombinant PDK4 protein [16]. Within the physiological context of intermittent feeding, loss of sensitivity of skeletal muscle PDK to suppression by pyruvate as a consequence of PDK4 upregulation may be geared to facilitate direction of glycolytically derived pyruvate toward lactate output rather than oxidation, with subsequent use for glucose synthesis [16]. Within the pathological context of prolonged starvation, trauma and sepsis, a further functional consequence of altered muscle PDK4 expression may relate to the fact that skeletal muscle protein, is relatively expendable to generate carbon skeletons of amino acids to act as additional gluconeogenic precursors. Skeletal muscle amino acids are transaminated with pyruvate to produce alanine, which is then released into the circulation. Increased expression of PDK4, which is less pyruvate sensitive (Figure 7), may permit the maintenance of adequately high pyruvate levels to ensure the removal of amine nitrogen from muscle in states 11 of negative nitrogen balance. Finally, diversion of pyruvate from oxidation toward lactate or alanine output by skeletal muscle may fuel excessive rates of endogenous glucose production and ultimately the development of hyperglycemia in insulin-resistant states. Thus pharmacological suppression of skeletal muscle PDK4 expression/activity may represent a potential strategy to oppose the development of hyperglycemia associated with insulin resistance [16,28,56]. In liver and kidney, PDK4 upregulation during starvation [3,41,62,63] may again participate in directing available pyruvate toward oxaloacetate formation, but in this case for entry into the gluconeogenic pathway and glucose synthesis. Concomitant upregulation of PDK2 may couple suppression of pyruvate oxidation with stimulation of pyruvate carboxylation via the common effector acetyl CoA. Conversely, impaired upregulation of PDK4 protein expression, resulting in an inappropriately high PDK2/PDK4 activity ratio, could result in increased flux via PDC at the expense of entry of pyruvate into gluconeogenesis. Support for this possibility comes from studies of PPAR-null mice where pharmacological inhibition of mitochondrial fatty acids oxidation results in severe hypoglycemia [64]. Regulatory properties of PDK It was reported that, only the PDK4 isoform prefers the lipoyl domain of the E3binding protein (E3BP) [65]. Such localization of PDKs on the swinging arms (Figure 1) and their transfer from one lipoyl domain to another gives the possibility of reaching almost all phosphorylation sites in the 30 molecules of E1 [56]. PDKs are activated via conformational changes when bound to the lipoyl domains [65,66]. Moreover, the activity of PDK is stimulated by the reduction and acetylation of the lipoyl residues occurring during PDC action [8]. Numerous studies with PDC isolated from various mammalian tissues indicate that, PDK is stimulated by NADH+H+ and acetyl CoA [1,67,68]. A high NADH+H+/NAD+ ratio causes reduction of the lipoyl groups through the reversal of the reaction catalyzed by dihydrolipoamide dehydrogenase subunit (E3), and a high acetyl CoA/CoA ratio promotes acetylation of the reduced lipoyl groups by dihydrolipoamide acetyltransferase subunit (E2) [69]. High NADH+H+ and acetyl CoA concentrations are found in mitochondria when fatty acid oxidation is intensive, for example during starvation, in high fat diet and diabetes [8,62]. Phosphorylation and concomitant inactivation of PDC by stimulated PDK is very typical for 12 such metabolic states. On the other hand, high NAD+ and CoA concentrations lead to PDK inhibition and the PDC remains active [11,67]. Pyruvate also exerts an inhibitory influence on PDK [70]. Isoform PDK2 is more sensitive to inhibition by pyruvate and stimulation by NADH+H+ and acetyl CoA than the other ones [8,70]. Competitive inhibition of PDK by ADP versus ATP was also considered to be essential in the regulation of the PDC [67,71]. Studies of PDK2 showed that, ADP dissociation from active sites is a limiting step in E2-activated catalysis by isoform 2. Conversion of all lipoyl groups in the E2-oligomer to the oxidized form greatly reduced the Michaelis constant (Km) of PDK2 for ATP [66]. The E1-component of mammalian PDC (pyruvate dehydrogenase subunit) has 3 phosphorylation sites. PDK isoforms show different activity during phosphorylation of the sites. Their relative activities toward site 1 are: PDK2 > PDK4 > PDK1 > PDK3; toward site 2: PDK3 > PDK4 > PDK2 > PDK1 [69]. In the case of site 3, the differences in the activity of the PDHK isoforms are not so clear. The PDK1 can phosphorylate all 3 sites, whereas PDK2, PDK3 and PDK4 each phosphorylates only sites 1 and 3 [28]. Earlier, it was reported that, only phosphorylation at sites 1 and 2 results in PDC inactivation, yet additional phosphorylation of site 3 inhibits reactivation of E1 by PDP [67]. However, according to new data, phosphorylation of either site leads to PDC inactivation, but in a different way [28,56]. For example, phosphorylation of site 1 of human PDC prevents its interaction with the substrate, pyruvate and a lipoyl domain, whereas modification of site 3 hinders E1 interaction with TDP [69]. Conversely, exogenous TDP added to the reaction medium results in inhibition of PDK activity [72,73]. TDP especially decreases the amount of phosphate that PDK1 incorporates in sites 2 and 3 and PDK2 in site 2 [28]. Tyrosine phosphorylation of PDK1 The tyrosine phosphorylation enhances PDK1 activity by promoting ATP and PDC binding. Functional PDC can form in mitochondria outside of the matrix in some cancer cells and PDK1 is commonly tyrosine phosphorylated in human cancers by diverse oncogenic tyrosine kinases localized to different mitochondrial compartments. Expression of phosphorylation-deficient, catalytic hypomorph PDK1 mutants in cancer cells leads to decreased cell proliferation under hypoxia and increased oxidative phosphorylation with enhanced mitochondrial utilization of pyruvate and reduced tumor growth in xenograft nude 13 mice. Together, tyrosine phosphorylation activates PDK1 to promote the Warburg effect and tumor growth [74]. Regulation of PDKs expression Besides the short-term regulation by the metabolites, a long-term control of PDK amount at the transcriptional level is very important. This aspect is presented thoroughly in some published reviews [8,69]. Regulation of PDK expression may occur at the levels of both mRNA and protein expression. The half-life of the PDK4 mRNA in Morris hepatoma cells is relatively short (~1.5 h) compared with that of PDK2 (t1⁄2 > 6 h) [75]. Increases in PDK2 mRNA and protein expression evoked by prolonged starvation on PDK isoform expression in liver and kidney are relatively similar (~2-fold) in magnitude, suggesting that regulation of PDK2 expression occurs primarily at the level of transcription [62]. Conversely, the relative increases in hepatic and renal PDK4 mRNA expression evoked by prolonged starvation (3.0 and 8.9 fold, respectively) are much higher than the corresponding increases in PDK4 protein expression (1.9 and 3.8 fold, respectively) [62]. Hence, both transcriptional and translational mechanisms are likely to participate in the long-term regulation of PDK4 expression in liver and kidney [62]. Interestingly, starvation increases PDK4 mRNA expression in brain and white adipose tissue without any corresponding increase in PDK4 protein expression, suggesting dominant regulation of PDK4 protein expression in these tissues at the level of translation. Insulin suppresses hepatoma PDK2 mRNA expression but is less effective in lowering hepatoma PDK4 mRNA expression [75]. Differential sensitivity of PDK2 and PDK4 to suppression by insulin may possibly be extrapolated to other tissues. Thus starvation increases only PDK4 in slow-twitch muscle but increases both PDK2 and PDK4 protein in fast-twitch skeletal muscle [40], which is relatively less insulin sensitive than slow-twitch skeletal muscle [76]. In cancer cells, the aberrant conversion of pyruvate into lactate instead of acetyl CoA in the presence of oxygen is known as the Warburg effect. The consequences and mechanisms of this metabolic peculiarity are incompletely understood. The p53 status is a key determinant of the Warburg effect. Wild-type p53 expression decreased levels of PDK2 and the product of its activity, the inactive form of the PDC, both of which are key regulators of pyruvate metabolism. Decreased levels of PDK2 and PDC in turn promoted conversion of pyruvate 14 into acetyl CoA instead of lactate. Thus, wild-type p53 limited lactate production in cancer cells unless PDK2 could be elevated. The wild-type p53 prevents manifestation of the Warburg effect by controlling PDK2 elucidating a new mechanism by which p53 suppresses tumorigenesis acting at the level of cancer cell metabolism [77]. Hypoxia induces PDK1 and PDK3 expression in cancer cells. Up-regulation of PDK1 and PDK3 under hypoxia induces metabolic switch and drug resistance. Given its unique feature in enzyme kinetics and biological function, PDK3 may represent the critical molecule that controls the metabolic switch and drug resistance in cancer cells, especially those with elevated HIF-1 levels. These findings provide novel insights about pathological processes of cancer cells and suggest that, PDK3 may be a new and pivotal target for tumor therapy [78,79]. PPARs are ligand-activated transcription factors that have hypolipidemic actions that control number of genes in several pathways of lipid metabolism [80]. PPAR, the predominant molecular target for the insulin-sensitizing thiazolidinediones, is most abundantly expressed in adipose tissue, where it induces adipocyte genes that increase fatty acids delivery (e.g. lipoprotein lipase) or uptake and sequestration (e.g. fatty acid transporter 1) [81]. Through its action to facilitate lipid trapping in adipose tissue, lipid delivery to tissues other than adipose tissue, including those express PDK4 at relatively high levels, is reduced. Use of PCR differential mRNA display, DNA microarrays and related techniques has shown that skeletal muscle PDK4 gene expression is suppressed by treatment of insulin-resistant rats with PPARagonists. This observation supports a direct inverse relationship between lipid entrapment in adipose tissue and tissue PDK4 expression [82]. PPAR and PPAR agonists were found to specifically up-regulate PDK4 mRNA expression, whereas PPAR activation selectively decreased PDK2 mRNA transcript abundance. Human muscle, hormonal and nutritional conditions may control PDK2 and PDK4 mRNA expression via a common signalling mechanism. In addition, PPARs appear to independently regulate specific PDK isoform transcript levels, which are likely to impart important metabolic mediation of fuel utilization by the muscle [83]. Previous studies have shown that starvation and experimental diabetes induce a stable increase in PDK activity in skeletal muscle [84-86], which explains the decreased activity of PDC and reduced glucose oxidation in these metabolic states. Skeletal muscle expresses 15 PDK2 and PDK4 isoforms in mammalian cells [34,35]. The increase in PDK activity with starvation and diabetes was shown to be largely due to a selective upregulation of PDK4 expression [30,37]. Because the plasma free fatty acids level increases with starvation and diabetes, free fatty acids has been suggested to stimulate PDK4 expression in skeletal muscle [37,87]. Free fatty acids is an endogenous ligand for the PPAR [88,89] and may increase PDK4 expression by stimulating PPAR [30,90]. Hyperinsulinemic euglycemic clamp resulted in decreases in PDK mRNA levels in human skeletal muscle, suggesting a direct effect of insulin on PDK expression [91]. The short exposure (i.e. 100 minutes) to insulin failed to demonstrate statistical significance for a small decrease in PDK4 mRNA level while demonstrating a significant decrease in PDK2 mRNA. Also, because plasma free fatty acids level falls during hyperinsulinemic glucose clamps, the possibility cannot be excluded that insulin decreased PDK expression indirectly by decreasing plasma free fatty acids (FFA). Insulin had a dramatic effect to suppress PDK4 (but not PDK2) mRNA expression in skeletal muscle independently of insulin effect to decrease plasma FFA levels. In addition and contrary to general belief, circulating FFA had little impact on PDK4 mRNA expression in skeletal muscle. The rapid and profound changes in PDK4 mRNA expression during fasting or re-feeding (or experimental diabetes) might be largely due to changes in circulating insulin rather than FFA [92]. Starvation and diabetes increase the level of PDK4 expression in most tissues [93]. Insulin treatment of diabetic rats reverses these changes [8]. Starvation also increases PDK2 expression in the liver and kidney [62]. Rapid increase in PDK4 mRNA expression by 3 fold and 14 fold was observed after 15 hours and 40 hours of fasting, respectively. PDC activity was significantly lowered after 40 hours of fasting, suggesting a PDK4-mediated increase in PDK protein and activity and a decrease in carbohydrate oxidation in resting skeletal muscle [94]. Increased expression of PDK4 mRNA and protein occurs in many tissues in response to starvation and chemical-induced diabetes [26,62]. Although less dramatic, starvation also increases PDK2 gene expression in the liver and kidney [27,62]. Conditions that affect expression of the other two known PDK isoforms, PDK1 and PDK3, have not been found [75] PDK isoforms are expressed differently in different cell types [16,95]. The expression of PDK1 is limited to heart and pancreas [30,37,95]. PDK2 is expressed with low amounts in 16 the lung and spleen [39], whereas PDK3 is found only in the testis, kidney and brain [39,95]. For example, the expression of PDK4 is found in the tissues that are important in the metabolic clearance of glucose such as liver and skeletal muscle [30,37,95]. This suggests that PDK4 is the critical isomer involved in the maintenance of glucose homeostasis. Consistent with this hypothesis, PDK4 is expressed in high levels during obesity and diabetes. In addition, an increase in PDK4 expression has been observed in response to starvation and insulin resistance [23,28]. Taking into account that PDK4 shows high specificity towards site 2 of PDC, the increase of PDK4 levels in response to starvation, insulin resistance and diabetes is believed to be important for site 2 phosphorylation. It is postulated that phosphorylation of site 2 by PDK4 results in limiting the rate of PDP-dependent reactivation of PDC, thus ensuring that, PDC remains in the inactive form. This postulate is supported by the observation that, reactivation of PDC in the heart, liver and skeletal muscle is much slower under conditions of high PDK4 expression than under conditions of low PDK4 expression [23]. Therefore, PDK4 seems to be an ideal target for long-term inhibition of PDK activity with respect to control of type 2 Diabetes mellitus as it was reported that, PDK4 deficiency lowers blood glucose and improves glucose tolerance in diet-induced obese mice [96]. Glucocorticoids, FFAs and insulin play important roles in setting the level of PDK expression. This level in turn determines the phosphorylation state and therefore the activity state of PDC. Thus the inactivation of PDC that occurs in most of the major tissues of the body during starvation and diabetes is likely explained by the effects that a decrease in insulin and the increase in glucocorticoids and FFAs have on PDK4 expression in these conditions. The increased expression of PDK4 in response to dexamethasone likely reflected a physiologically important effect of glucocorticoids. It occurred in the intact liver in response to administration of dexamethasone and probably in other tissues as well, given that glucocorticoid receptors are present in many cells of the body [75]. The induction of PDK4 expression by the increase in blood levels of glucocorticoids during starvation promotes gluconeogenesis by blocking irreversible loss of pyruvate carbon to the tricarboxylic acid cycle. For the same reason, overexpression of PDK4 in response to excess glucocorticoids very likely contributes to diabetes induced by excessive therapeutic use of steroids and comorbid with Cushing syndrome [97]. 17 The FoxOs class of transcription factors are believed to be a central metabolic regulator in glucose and lipid metabolism [98]. Glucocorticoids increase PDK4 mRNA and thereby protein expression, which phosphorylates PDC, thereby preventing the formed pyruvate from undergoing mitochondrial oxidation. This increase in PDK4 expression is mediated by the presence of FoxOs in the nucleus. Glucocorticoids promoted nuclear retention of FoxO1 through a decrease in phosphorylation of heat shock proteins (hsps). Manipulation of FoxO1 through agents that interfere with its nuclear shuttling or acetylation were effective in reducing dexamethasone-induced increase in PDK4 protein expression. Manipulating FoxO could assist in devising new therapeutic strategies to optimize cardiac metabolism and to prevent PDK4-induced cardiac complications [99]. Cardiac metabolism of glucose is very tightly controlled in order to maintain the variable energy demand that is required by cardiac tissue. Energy metabolism of the cardiac myocyte can be regulated within seconds up to a few minutes or chronically regulated within the time frame of hours to days. Glucose metabolism is activated in early myocardial ischemia and in response to an increased need of high-energy phosphate in the healthy heart during extreme physical activity. In myocardial ischemia, inhibition of PDK expression would be beneficial in order to shift the myocardial metabolism from the adult towards the fetal phenotype, thus metabolising more glucose than fat to preserve myocardial integrity. In particular, PDK4 is significantly less expressed under nebivolol both during O2 perfusion and simulated ischemia, an effect practically negligible under atenolol [100]. The peroxisome proliferator-activated receptor gamma (PPAR- coactivator-1 alpha (PGC-1) stimulates the expression of PDK4. PGC-1 belongs to a family of transcriptional coactivators that includes PGC-1 and the PGC-1-related coactivator. PGC-1 stimulates hepatic gluconeogenesis and expression of the phosphoenolpyruvate carboxykinase and glucose-6-phosphatase genes. The ERR and ERR stimulate the PDK4 gene in hepatoma cells, suggesting a novel role for ERRs in controlling pyruvate metabolism. In addition, both ERR isoforms recruit PGC-1 to the PDK4 promoter. Insulin, which decreases the expression of the PDK4 gene, inhibits the induction of PDK4 by ERR and ERR. The FoxO1 binds the PDK4 gene and contributes to the induction of PDK4 by ERRs and PGC-1. Insulin suppresses PDK4 expression in part through the dissociation of FoxO1 and PGC-1 from the 18 PDK4 promoter. These data demonstrates a key role for the ERRs in the induction of hepatic PDK4 gene expression [101]. PDK inhibitors Currently, the most commonly known inhibitor of PDK that has reached clinical trial stage is dichloroacetate (DCA). Recently, other inhibitors of PDK activity such as triterpenes [102], analogues of ATP, AZD7545 and lactones and halogenated acetophane [103] have been identified (Figure 8). Selective inactivation of PDK isoforms by the specific inhibitor DCA has been shown to de-repress a mitochondrial potassium-ion channel axis, trigger apoptosis in cancer cells and inhibit tumor growth [103]. DCA has been shown to have beneficial effects in diabetes, lactic acidosis and myocardial ischemia [104]. Biochemical studies indicated that DCA and ADP synergistically inhibit activities of PDK isoforms [66]. Recent mutational analyses and the PDK2 structure in complex with DCA revealed that DCA binds to the N-terminal domain of PDK2 [105]. However, the inhibition mechanism of DCA is not known, as no structural change in PDK2 was observed upon DCA binding. The glucose-lowering compound AZD7545, manufactured by AstraZeneca®, UK, has been shown to inhibit PDK2, resulting in enhanced glucose breakdown through PDC flux in tissues from diabetic animals [106]. The antitumor compound radicicol (Figure 8) inhibits the activities of Hsp90 and Topo VI by blocking ATP binding to these enzymes [55]. PDKs is also inhibited by radicicol [107]. The crystal structures of the human PDK1-AZD7545, PDK1-DCA and PDK3-L2- radicicol complexes have been elucidated [14]. Mechanism of DCA as a PDK inhibitor DCA (Figure 8) is a non-hydrolysable analogue of pyruvate that has been shown to improve the diabetic condition [108,109]. DCA has been used for other conditions in which PDC activity is reduced such as ischemia [110] and cardiac insufficiency [104]. DCA binds to PDK at an allosteric site that was mapped using the crystal structure of human PDK2 complexed to DCA [111]. This site is located in the N-terminal domain and is formed by residues Leu 53, Tyr 80, Ser 83, Ile 111, Arg 112, His 115, Ser 153, Arg 154, Ile 157, Arg 158 and Ile161 [111] (Figure 9). Most of these residues are conserved in all known PDKs isoforms. The carboxylate group of DCA forms a salt-bridge with Arg158 while the chlorine is buried in the hydrophobic portion of the DCA binding site [111]. It is believed that, binding 19 of DCA to PDK prevents dissociation of ADP from the active site of PDK, thus locking PDK in its inactive form [66]. Clinical applications of PDK inhibitors PDK has received attention as a target for type 2 Diabetes mellitus therapy due to its role in glucose metabolism and diabetes. Therefore, there have been a number of efforts to identify and design PDK inhibitors [112]. DCA has proved not to be ideal for long term treatment of type 2 Diabetes mellitus due to its toxicity [109,113,114]. The toxicity of DCA results from it being metabolized to glyoxylate which is converted to oxalate. High amounts of oxalate lead to abnormal oxalate metabolism [109,115]. Furthermore, DCA stimulates thiamine-dependent enzymes which cause depletion of thiamine in the body as thiamine have a role in activating the PDKs [109]. It is, therefore, imperative to investigate other useful drugs that can inhibit PDK activity for the therapy of diabetes, insulin resistant diabetes and other metabolic disorders. DCA was used orally in the treatment of children with congenital lactic acidosis caused by mutations in the PDC. The case histories of 46 subjects were analyzed with regard to diagnosis, clinical presentation and response to DCA. DCA decreased blood and cerebrospinal fluid lactate concentrations and was generally well tolerated. DCA may be particularly effective in children with PDC deficiency by stimulating residual enzyme activity and consequently, cellular energy metabolism. A controlled trial is needed to determine the definitive role of DCA in the management of this devastating disease [116]. Hyperthyroidism increases heart rate, contractility, cardiac output and metabolic rate. It is also accompanied by alterations in the regulation of cardiac substrate use. Specifically, hyperthyroidism increases the ex vivo activity of PDK, thereby inhibiting glucose oxidation via PDC. Cardiac hypertrophy is another effect of hyperthyroidism, with an increase in the abundance of mitochondria. Although the hypertrophy is initially beneficial, it can eventually lead to heart failure. The inhibition of glucose oxidation in the hyperthyroid heart in vivo is mediated by PDK. Relieving this inhibition using PDK inhibitors can increase the metabolic flexibility of the hyperthyroid heart and reduce the level of hypertrophy that develops while 20 maintaining the increased cardiac output required to meet the higher systemic metabolic demand [117]. Cancer cells have inactivated PDC; therefore, inhibition of PDK is suggested as a possible line of treatment for cancer. Recent progress has shown that apoptosis in solid tumor cells can be therapeutically invoked by the forcing a shift in metabolic energy production from aerobic glycolysis in cancer cells to a glucose oxidation pathway of healthy normal functioning cells by inhibiting mitochondrial PDK [118]. By activating the TCA, an inhibition of PDK subsequently decreases proliferation and inhibits tumor growth, without apparent toxicity in vivo. Inhibition of PDK may be an appropriate therapy for patients with metastatic breast cancer. DCA has been shown to reverse impaired oxidative metabolism of pyruvate in humans (specifically in children) for treatment of lactic acidosis for over 30 years [119]. Conclusions PDC catalyzes the irreversible oxidative decarboxylation of pyruvate into acetyl CoA. The control of PDC reaction is crucial to avoid depletion of carbohydrate derived carbon skeletons which cannot be resynthesised by the body [1]. Prolonged fasting or high fat diets inactivate the PDC in order to minimize loss of pyruvate carbon to conserve it for maintaining the blood glucose levels. PDK inactivates PDC by phosphorylation [2]. This property highlights a big area of scientific research in order to establish several PDKs inhibitors taking the advantage of treating several diseases such as hormonal abnormalities, heart diseases, cancer and other immunological and chronic diseases in the near future to improve the main stream of clinical practice. References [1] Yeaman SJ (1989) The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J 257: 625- 632. [2] Randle PJ (1995) Metabolic fuel selection- general investigation at the whole body level. Proc Nutr Soc 54: 317-27. [3] Sugden MC, Bulmer K, Holness MJ (2001) Fuel-sensing mechanisms integrating lipid and carbohydrate utilization. Biochem Soc Trans 29: 272–278. 21 [4] Schnaitman C, Greenawalt JW (1968) Enzymatic properties of the inner and outer membranes of rat liver mitochondria. J Cellu Biol 38: 158-175. [5] Addink A, Boer P, Wakabayashi T, Green D (1972) Enzyme localisation in beef-heart mitochondria. A biochemical and electronmicroscope study. Eur J Biochem 29: 4759. [6] Patel MS, Roche TE (1990) Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J 4: 3224-3233. [7] Cate RL, Roche TE, Davis LC (1980) Rapid intersite transfer of acetyl groups and movement of pyruvate dehydrogenase component in the kidney pyruvate dehydrogenase complex. J Biol Chem 225: 7556-7562. [8] Harris RA, Bowker-Kinley MM, Huang B, Wu P (2002) Regulation of the activity of the pyruvate dehydrogenase complex. Adv Enzyme Regul 42: 249-259. [9] Strumiło S, Czygier M, Kondracikowska J, Dobrzyn P, Czerniecki J (2002) Kinetic and spectral investigation of allosteric interaction of coenzymes with 2-oxo acid dehydrogenase complexes. J Mol Stru 614: 221–226. [10] Strumiło S (2005) Short-term regulation of the mammalian pyruvate dehydrogenase complex. Acta Biochimica Polonica 52: 759-64. [11] Sugden MC, Holness MJ (2003) Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab 284: E855-862. [12] Patel MS, Korotchkina LG (2001) Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med 4: 191-197. [13] Pratt ML, Roche TE (1979) Mechanism of pyruvate inhibition of kidney pyruvate dehydrogenase kinase and inhibitory by pyruvate and ADP. J Biol Chem 254: 71917196. [14] Kato M, Chuang JL, Tso S, Wynn RM, Chuang DT (2005) Crystal structure of pyruvate dehydrogenase kinase bound to lipol domain 2 of human pyruvate dehydrogenase complex. EMBO J 24: 1763-1774. [15] Steussy CN, Popov KM, Bowker-Kinley MM, Soan RB, Harris RA, Hamilton JA (2001) Structure of pyruvate dehydrogenase kinase: Novel folding pattern for a seine 22 protein kinase. J Biol Chem 276: 37443-37450. [16] Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM (1998) Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J 329: 191-196. [17] Hucho F, Randall DD, Roche TE, Burgett MW, Pelley JW, Reed LJ (1972) α Keto acid dehydrogenase complexes. XVII. Kinetic and regulatory properties of pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphotase from bovine kidney and heart. Arch Biochem Biophys 151: 328-340. [18] Yeaman SJ, Hutcheson ET, Roche TE, Pettit FG, Brown JR, Reed LJ, Watson DC, Dixon GH (1978) Sites of phosphorylation on pyruvate dehydrogenase from bovine kidney and heart. Biochem 17: 2364-2370. [19] Teague WM, Pettit FH, Yeaman SJ, Reed LJ (1979) Function of phosphorylation sites on pyruvate dehydrogenase. Biochem Biophys Res Commun 87: 651-657. [20] Sugden MC, Holness MJ (1989) Effects of re-feeding after prolonged starvation on pyruvate dehydrogenase activities in heart, diaphragm and selected skeletal muscles of the rat. Biochem J 262: 669–672. [21] Sale GJ, Randle PJ (1981) Analysis of site occupancies in [32P] phosphorylated pyruvate dehydrogenase complexes by aspartyl- prolyl cleavage of tryptic phosphopeptides. Eur J Biochem 120: 535–540. [22] Korotchkina LG, Patel MS (1995) Mutagenesis studies of the phosphorylation sites of recombinant human pyruvate dehydrogenase. Site-specific regulation. J Biol Chem 270: 14297−14304. [23] Holness MJ, Sugden MC (1989) Pyruvate dehydrogenase activities during the fed-to starved transition and on re-feeding after acute or prolonged starvation. Biochem J 258: 529–533. [24] Dahl HH, Hunt SM, Huttchison WM, Brown GK (1987) The human pyruvate dehydrogenase complex. Isolation of cDNA clones for the E1 alpha subunit, sequence analysis, and characterisation of the mRNA. J Biol Chem 15: 7398-7403. [25] Randle PJ (1986) Fuel sensing in animals. Biochem Soc Trans 14: 799-806. 23 [26] Wu P, Sato J, Zhao Y, Jaskiewicz J, Popov KM, Harris RA (1998) Starvation and diabetes increase the amount of pyruvate dehydrogenase kinase isoenzyme 4 in rat heart. Biochem J 329: 197-201. [27] Sugden MC, Fryer LG, Orfali KA, Priestman DA, Donald E, Holness MJ (1998) Studies of the long-term regulation of hepatic pyruvate dehydrogenase kinase. Biochem J 329: 89-94. [28] Kolobova E, Tuganova A, Boulatnikov I, Popov KM (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J 358: 69077-69082. [29] Holness MJ, Kraus A, Harris RA, Sugden MC (2000) Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes 49: 775–781. [30] Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA (1999) Mechanism responsible for the inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes 48: 1593-1599. [31] Patel MS, Korotchkina LG (2006) Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans 34: 217-222. [32] Popov KM, Kedishvili NY, Zhao Y, Shimomura Y, Crabb DW, Harris RA (1993) Primary structure of pyruvate dehydrogenase kinase establishes a new family of eukaryotic protein kinases. J Biol Chem 268: 26602–26606. [33] Popov KM, Kedishvili NY, Zhao Y, Gudi R, Harris RA (1994) Molecular cloning of the p45 subunit of pyruvate dehydrogenase kinase. J Biol Chem 269: 29720-29724. [34] Gudi R, Bowker-Kinley MM, Kedishvili NY, Zhao Y, Popov KM (1995) Diversity of the pyruvate dehydrogenase kinase gene family in humans. J Biol Chem 270: 28989– 28994. [35] Rowles J, Scherer SW, Xi T, Majer M, Nickle DC, Rommens JM, Popov KM, Harris RA, Riebow NL, Xia J, Tsui LC, Bogardus C, Prochazka M (1996) Cloning and characterization of PDK4 on 7q21.3 encoding a fourth pyruvate dehydrogenase kinase isoenzyme in human. J Biol Chem 271: 22376–22382. 24 [36] Sugden MC, Kraus A, Harris RA, Holness M.J (2000) Fibretype specific modification of the activity and regulation of skeletal muscle pyruvate dehydrogenase kinase (PDK) by prolonged starvation and refeeding is associated with targeted regulation of PDK isoenzyme 4 expression. Biochem J 346: 651–657. [37] Sugden MC, Bulmer K, Gibbons GF, Holness MJ (2001) Role of peroxisome proliferator-activated receptor-alpha in the mechanism underlying changes in renal pyruvate dehydrogenase kinase isoform 4 protein expression in starvation and after refeeding. Arch Biochem Biophys 395: 246–252. [38] Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJ, Spriet LL (2001) Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab 281: E1151-8. [39] Huang B, Gudi R, Wu P, Harris RA, Hamilton J, Popov KM (1998) Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J Biol Chem 273: 17680-17688. [40] Sugden MC, Kraus A, Harris RA, Holness MJ (2000) Expression and regulation of pyruvate dehydrogenase kinase isoforms in the developing rat heart and in adulthood: role of thyroid hormone status and lipid supply. Biochem J 346: 651-657. [41] Sugden MC, Bulmer K, Gibbons GF, Knight BL, Holness MJ (2002) Peroxisomeproliferator activated receptor (PPAR) deficiency leads to dysregulation of hepatic lipid and carbohydrate metabolism by fatty acids and insulin. Biochem J 364: 361368. [42] Davier JR, Wynn RM, Meng M, Huang YS, Aalund G, Chaung DT, Lau KS (1995) Expression and characterisation of branched-chain alpha-ketoacid dehydrogenase kinase from rat. Is it a histidine kinase?. J Biol Chem 270: 19861-19867. [43] Harris RA, Popov KM, Zhao Y, Kedishvilli NY, Shimomura Y, Crabb DW (1995) A new family of protein kinases-The mitochondrial protein kinases. Advan Enzyme Regul 35: 147-162. [44] Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596. [45] Kwon HS, Harris RA (2004) Mechanisms responsible for regulation of pyruvate dehydrogenase kinase 4 gene expression. Adv Enzyme Regul 44: 109-121. 25 [46] Rossman MG, Moras D, Olsen KW (1974) Chemical and biological evolution of nucleotide-binding protein. Nature 250: 194-199. [47] Alex LA, Simon MI (1994) Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genetics 10: 133-138. [48] Bergerat A, deMassy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414-417. [49] Machius M, Chuang JL, Wynn RM, Tomchick DR, Chuang DT (2001) Structure of rat BCKD kinase: nucleotide-induced domain communication in a mitochondrial protein kinase. Proc Natl Acad Sci USA 98: 11218-11223. [50] Thelen JJ, Miernyk JA, Randall DD (2000) Pyruvate dehydrogenase kinase from Arabidopsis thaliana: a protein histidine kinase that phosphorylates serine residues. Biochem J 349: 195–201. [51] Mooney BP, David NR, Thelen JJ, Miernyk JA, Randall DD (2000) Histidine Modifying Agents Abolish Pyruvate Dehydrogenase Kinase Activity. Biochem Biophys Res Commun 267: 500-504. [52] Tuganova A, Yoder MD, Popov KM (2001) An essential role of Glu-243 and His-239 in the phosphotransfer reaction catalyzed by pyruvate dehydrogenase kinase. J Biol Chem 276: 17994-17999. [53] Tovar-Mendez A, Hirani TA, Miernyk JA, Randall DD (2005) Analysis of the catalytic mechanism of pyruvate dehydrogenase kinase. Arch Biochem Biophys 434: 159-168. [54] Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI (2001) Nucleotide binding by the histidine kinase CheA. Nature Stru Biol 8: 353-360. [55] Corbett KD, Berger JM (2003) Structure of the topoisomeraseVI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J 22: 151163. [56] Korotchkina LG, Patel MS (2001) Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem 276: 37223-37229. 26 [57] Cooper RH, Randle PJ, Denton RM (1975) Stimulation of phosphorylation and inactivation of pyruvate dehydrogenase by physiological inhibitors of the pyruvate dehydrogenase reaction. Nature 275: 808-809. [58] Czygier MB, Strumiło SA (1995) Changes in some properties of the aurochs heart pyruvate dehydrogenase complex induced by its partial phosphorylation. Ukr Biokhim Zh 67: 25–28. [59] Yang D, Gong X, Yakhnin A, Roche TE (1998) Requirements for the adaptor protein role of dihydrolipoyl acetyltransferase in the up-regulated function of the pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphatase. J Biol Chem 273: 14130-14137. [60] Priestman DA, Orfali KA, Sugden MC (1996) Pyruvate inhibition of pyruvate dehydrogenase kinase. Effects of progressive starvation and hyperthyroidism in vivo, and of dibutyryl cyclic AMP and fatty acids in cultured cardiac myocytes. FEBS Lett 393: 174-178. [61] Holness MJ, Bulmer K, Gibbons GF, Sugden MC (2002) Up-regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) protein expression in oxidative skeletal muscle does not require the obligatory participation of peroxisome-proliferatoractivated receptor alpha (PPARalpha). Biochem J 366: 839–846. [62] Wu P, Blair PV, Sato J, Jaskiewicz J, Popov KM, Harris RA (2000) Starvation increases the amount of pyruvate dehydrogenase kinase in several mammalian tissues. Arch Biochem Biophys 381: 1–7. [63] Wu P, Peters JM, Harris RA (2001) Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor alpha. Biochem Biophys Res Commun 287: 391-396. [64] Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP (1998) A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferatoractivated receptor alpha-deficient mice. J Clin Investig 102: 1083-1091. [65] Roche TE, Hiromasa Y, Turkan A, Gong X, Peng T, Yan X, Kasten SA, Bao H, Dong J (2003) Essential roles of lipoyl domains in the activated function and control 27 of pyruvate dehydrogenase kinases and phosphatase isoform 1. Eur J Biochem 270: 1050–1056. [66] Bao H, Kasten SA, Yan X, Roche TE (2004) Pyruvate dehydrogenase kinase isoform 2 limited and further inhibited by slowing down the rate of dissociation of ADP. Biochem 43: 13432–13441. [67] Wieland OH (1983) The mammalian pyruvate dehydrogenase complex: structure and regulation. Reviews of Physiology, Biochem & Pharmacol 96: 123-170. [68] Reed LJ (2001) A trail of research from lipoic acid to α- keto acid dehydrogenase complexes. J Biol Chem 276: 38329–38336. [69] Patel MS, Korotchkina LG (2003) The biochemistry of the pyruvate dehydrogenase complex. Biochem Mol Biol Education 31: 5-15. [70] Bao H, Kasten SA, Yan X, Hiromasa Y, Roche TE (2004) Pyruvate dehydrogenase kinase isoform 2 activity stimulated by speeding up the rate of dissociation of ADP. Biochem 43: 13442–13451. [71] Strumiło SA, Senkevich SB, Vinogradov VV (1981) Characterization of kinase activity of pyruvate dehydrogenase complex from bovine adrenals. Biokhimiya 46: 974-978 (English edition as Biochemistry- Moscow 46: 788-792). [72] Walsh DA, Cooper RH, Denton RM, Bridges BJ, Randle PJ (1976) The elementary reactions of the pig heart pyruvate dehydrogenase complex. A study of the inhibition by phosphorylation. Biochem J 157: 41–67. [73] Czygier M, Strumiło S (1995) An investigation of the kinase activity of aurochs heart pyruvate dehydrogenase complex. Biochem Mol Biol Inter 37: 313-317. [74] Hitosugi T, Fan J, Chung T-W, Lythgoen K, Wang X, Xie J, Ge Q, Gu T-L, Polakiewicz R, Roessel J, Chen G, Boggon T, Lonial S, Fu H, Khuri F, Kang S, Chen J (2011) Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 44(6): 864-877. [75] Huang B, Wu P, Bowker-Kinley MM, Harris RA (2002) Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor- ligands, glucocorticoids, and insulin. Diabetes 51: 276–283, 2002. [76] James DE, Jenkins AB, Kraegen EW (1985) Heterogeneity of insulin action in 28 individual muscles in vivo: euglycemic clamp studies in rats. Am J Physiol Endocrinol Metab 248: E567–E574. [77] Contractor T, Harris C (2012) p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res 72: 560-567. [78] Kim J, Tchernyshyov I, Semenza G, Dang CV (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3(3): 177-185. [79] Lu C-W, Lin S-C, Chen K-F, Lai Y-Y, Tsai S-J (2008) Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem 283 (42): 28106-28114. [80] Ferrè P (2004) The biology of peroxisome proliferator-activated receptors relationship with lipid metabolism and insulin sensitivity. Diabetes 53: S43-S50. [81] Kersten S, Desvergne B, Wahli W (2000) Roles of PPARs in health and disease. Nature 405: 421-424. [82] Way JM, Harrington WW, Brown KK, Gottschalk WK, Sundseth SS, Mansfield TA, Ramachandran RK, Willson TM, Kliewer SA (2001) Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor gamma activation has coordinate effects on gene expression in multiple insulinsensitive tissues. Endocrinol 142: 1269–1277. [83] Abbot E, McCormack J, Reynet C, Hassall D, Buchan K, Yeaman S (2005) Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS J 272: 3004-3014. [84] Fuller SJ, Randle PJ (1984) Reversible phosphorylation of pyruvate dehydrogenase in rat skeletal-muscle mitochondria. Effects of starvation and diabetes. Biochem J 219: 635–646. [85] Stace PB, Fatania HR, Jackson A, Kerbey AL, Randle PJ (1992) Cyclic AMP and free fatty acids in the longer-term regulation of pyruvate dehydrogenase kinase in rat soleus muscle. Biochim Biophys Acta 1135: 201–206. [86] Feldhoff PW, Arnold J, Oesterling B, Vary TC (1993) Insulin-induced activation of pyruvate dehydrogenase complex in skeletal muscle of diabetic rats. Metabolism 42: 29 615-623. [87] Randle PJ, Priestman DA, Mistry S, Halsall A (1994) Mechanisms modifying glucose oxidation in diabetes mellitus. Diabetologia 37 Suppl 2: S155–S161. [88] Gearing KL, Gottlicher M, Widmark E, Banner CD, Tollet P, Stromstedt M, Rafter JJ, Berge RK, Gustafsson JA (1994) Fatty acid activation of the peroxisome proliferator activated receptor, a member of the nuclear receptor gene superfamily. J Nutr 124, Suppl 8: 1284S-1288S. [89] Forman BM, Chen J, Evans RM (1997) Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator activated receptors alpha and delta. Proc Natl Acad Sci USA 94: 4312–4317. [90] Holness MJ, Smith ND, Bulmer K, Hopkins T, Gibbons GF, Sugden MC (2002) Evaluation of the role of peroxisome proliferator activated receptor alpha in the regulation of cardiac pyruvate dehydrogenase kinase 4 protein expression in response to starvation, high-fat feeding and hyperthyroidism. Biochem J 364: 687–694. [91] Majer M, Popov KM, Harris RA, Bogardus C, Prochazka M (1998) Insulin downregulates pyruvate dehydrogenase kinase (PDK) mRNA: potential mechanism contributing to increased lipid oxidation in insulin-resistant subjects. Mol Genet Metab 65: 181-186. [92] Lee F, Zhang L, Zheng D, Choi W, Youn J (2004) Insulin suppresses PDK-4 expression in skeletal muscle independently of plasma FFA. Am J Physiol Endocrinol Metab 287: E69-E74. [93] Harris RA, Huang B, Wu P (2001) Control of pyruvate dehydrogenase kinase gene expression. Adv Enzyme Regul 41: 269-288. [94] Spriet LL, Tunstall RJ, Watt MJ, Mehan KA, Hargreaves M, Cameron-Smith D (2004) Pyruvate dehydrogenase activation and kinase expression in human skeletal muscle during fasting. J App Physiol 96: 2082-2087. [95] Bowker-Kinley MM, Popov KM (1999) Evidence that pyruvate dehydrogenase kinase belongs to the ATPase/kinase superfamily. Biochem J 344: 47-53. [96] Jeoung N, Harris R (2008) Pyruvate dehydrogenase kinase-4 deficiency lowers blood 30 glucose and improves glucose tolerance in diet-induced obese mice Am J Physiol Endocrinol Metab 295: E46–E54, 2008. [97] Orth DN (1995) Cushing’s syndrome. N Engl J Med 332: 791–803. [98] Nakae J, Oki M, Cao Y (2008) The FOXO transcription factors and metabolic regulation. FEBS Lett 582: 54-67. [99] Puthanveetil P, Wang Y, Wang F, Kim M, Abrahani A, Rodrigues B (2010) The increase in cardiac pyruvate dehydrogenase kinase-4 after short-term dexamethasone is controlled by an Akt-p38-forkhead box other factor-1 signaling axis. Endocrinol 151(5): 2306-2318. [100] Holzwart E, Gasser S, Roessl U, Buehner A, Ablasser K, Friehs I, Pieske B., Mächler H, Yates A, Tscheliessnig KH, Mangge H, Porta S, Gasser R (2010) Effect of betablockers on the regulation of PDK (pyruvate dehydrogenase kinase) gene expression in both normoxic and hypoxic myocardium. J Clin Basic Cardiol 13(1-4): 12-18. [101] Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard G, Wang F, McDonnell D, Unterman, T, Elam M, Park E (2006) Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 Gene Expression. J Biol Chem 281 (52): 398973990. [102] Aicher TD, Damon RE, Koletar J, Vinluan CC, Brand LJ, Gao J, Shetty SC, Kaplan EL, Mann WR (1999) Triterpene and diterpene inhibitors of pyruvate dehydrogenase kinase. Bioorganic Medicinal Chemistry Lett 9: 2223-2228. [103] Mann RW, Dragland JC, Vinluan CC, Vedananda PA, Bell PA, Aicher TD (2000) Diverse mechanisms of inhibition of pyruvate dehydrogenase kinase by structurally distinct inhibitors. Biochim Biophys Acta 1480: 283-292. [104] Bersin RM, Stacpoole PW (1997) Dichloroacetate as metabolic therapy for myocardial ischemia and failure. Am Heart J 134: 841-855. [105] Klyuyeva A, Tuganova A, Popov KM (2007) Amino acid residues responsible for the recognition of dichloroacetate by pyruvate dehydrogenase kinase 2. FEBS Lett 581: 2988–2992. 31 [106] Mayers RM, Leighton B, Kilgour E (2005) PDH kinase inhibitors: a novel therapy for type II diabetes?. Biochem Soc Trans 33: 367–370. [107] Besant PG, Lasker MW, Bui CD, Turck CW (2002) Inhibition of branched-chain αketo acid dehydrogenase kinase and Sln1 yeast histidine kinase by the antifungal antibiotic radicicol. Mol Pharmacol 62: 289-296. [108] Whitehouse S, Cooper RH, Randle PJ (1974) Mechanism of activation of pyruvate dehydrogenase by dichloroacetate and other halogenated carboxylic acids. Biochem J 141: 761-777. [109] Stacpoole PW, Moore GW, Kornhauser DM (1979) Toxicity of chronic dichloroacetate. New Eng J Med 300: 372-376. [110] Preiser JC, Moulart D, Vincent JL (1990) Dichloroacetate administration in the treatment of endotoxic shock. Circ Shock 30: 221-228. [111] Knoechel TR, Tucker AD, Robinson CM, Philips C, Wendy T, Bungay PJ, Kasten SA, Roche TE, Brown DG (2006) Regulatory roles of the N-terminal based on crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligand. Biochem 45: 402-415. [112] Kakkar R (2011) Structure-Based Design of PDHK2 Inhibitors from Docking Studies. Intl Res J Pharmaceu 10 (2): 51-59. [113] Katz R, Tai CN, Diener RM, McConnell RF, Semonick DE (1981) Dichloroacetate sodium: 3-month oral toxicity studies in rats and dogs. Toxicol Appl Pharmacol 57: 273-287. [114] Toth GP, Kelty KC, George E, Read EJ, Smith MK (1992) Adverse male reproductive effects following subchronic exposure of rats to sodium dichloroacetate. Fundam App Toxicol 19: 57-63. [115] Yount EA, Felten SY, O’Connor BL, Eterson RG, Powel RS, Yum MN, Harris RA (1982) Comparison of the metabolic and toxic effects of 2-choloropropionate and dichloroacetate. J Pharmacol Exp Ther 222: 501-508. [116] Berendzen K, Theriaque D, Shuster J, Stacpoole P (2006) Therapeutic potential of dichloroacetate for pyruvate dehydrogenase complex deficiency. Mitochondrion 6 (3): 126-135. 32 [117] Atherton H, Dodd M, Heather L, Schroeder M, Griffin J, Radda G, Clarke K, Tyler D (2011) Role of Pyruvate Dehydrogenase Inhibition in the Development of Hypertrophy in the Hyperthyroid Rat Heart: A Combined Magnetic Resonance Imaging and Hyperpolarized Magnetic Resonance Spectroscopy Study. Circu 123: 2552-2561. [118] Gillies RJ, Gatenby RA (2007) Adaptive landscapes and emergent phenotypes why do cancers have high glycolysis?. J Bioenerg Biomembr 39(3): 251-257. [119] Spechler SJ, Esposito AL, Koff RS, Hong WK (1978) Lactic acidosis in oat cell carcinoma with extensive hepatic metastases. Arch Intern Med 138(11): 1663-1664. 33 Figure 1. Scheme of the mammalian pyruvate dehydrogenase complex structure. Only 2 subunits of E2 with E1 and E3 components are shown. ID: Inner domain; BD: Binding domain; LD: Lipoyl domain; TDP: Thiamine diphosphate; FAD: Flavin adenine dinucleotide (oxidised form); PDK: Pyruvate dehydrogenase kinase; PDP: Pyruvate dehydrogenase phosphatase [10]. 34 Figure 2. The five sequential reactions catalysed by the pyruvate dehydrogenase complex for oxidative decarboxylation of pyruvate to acetyl CoA. TPP: Thiamine pyrophosphate; Lip: Lipoic acid; FADH2: Flavin adenine dinucleotide (reduced form); FAD: Flavin adenine dinucleotide (oxidised form); NADH+H+: Nicotinamide adenine dinucleotide (reduced form); NAD: Nicotinamide adenine dinucleotide (oxidised form) [10] 35 Figure 3: Regulation of PDC activity by a phosphorylation-dephosphorylation cycle [17-19]. 36 Figure 4: The long-term and short-term regulation of PDK. The reaction catalysed by PDC is also shown. The arrows show factors that induce PDK expression and activity. The broken lines indicate factors that inhibit PDK expression and activity [14]. 37 Figure 5. Schematic of the reactions controlling pyruvate dehydrogenase complex [3]. 38 PDK1 PDK2 PDK3 PDK4 Figure 6. Ribbon representation of the structure of PDK isoenzymes as retrieved from the National Center of Bioinformatics-USA (NCBI). 39 Figure 7. Comparison of kinetic parameters of mammalian PDK1, PDK2 and PDK4. Thicknesses of the lines indicate the relative activities of individual PDK isoforms toward the phosphorylation sites on the PDC- subunit (E1) [16,56]. 40 Radicicol Sodium dichloroacetate Triterpine Halogenated acetophane Non-hydrolysable ATP analogue adenosine5,-[β,γ-imido]triphosphate (p[NH]ppA Lactones (Dehydrobietylamine) AZD7545 Figure 8: Structures of some PDK inhibitors [103]. 41 Figure 9: Interactions between DCA and the residues in the DCA binding site of PDK2. The DCA allosteric pocket in PDK is coloured where red indicates region with negative charge, blue a positive charge region and white a neutral region. The DCA molecule is coloured cyan. The red and yellow rings in DCA molecule denote the oxygen and chlorine atoms, respectively. The green dotted line shows hydrogen bonds between DCA and Tyr 80 while the solid grey line represents a salt bridge that is formed between Arg 145 and DCA [111]. 42 Table 1. General data about PDK isoenzymes (EC number 2.7.11.2) as retrieved from the NCBI genebank databases. PDK isoenzyme PDK1 PDK2 PDK3 PDK4 1372 2511 1472 1798 AK312700.1 NM001199899.1 BC015948.1 U54617.1 Entrez code 5163 5164 5165 5166 Uniport code Q15118 Q15119 Q15120 Q16654 Parameter Gene Size (bp) NCBI Accession no. Ref. Seq. (mRNA) NM002610.3 NM001199898.1 NM001142386.2 NM002612.3 Ref. Seq. (protein) NP002601.1 NP001186827.1 NP001135858.1 NP002603.1 Chromosome 2 Chromosome 17 Chromosome X Chromosome 7 Gene Location 43