* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download chapt09_lecture
Nucleic acid tertiary structure wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Oncogenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Human genome wikipedia , lookup
DNA polymerase wikipedia , lookup
Minimal genome wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
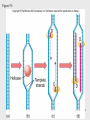
History of RNA biology wikipedia , lookup
DNA vaccination wikipedia , lookup
Epigenomics wikipedia , lookup
Frameshift mutation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
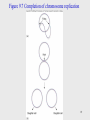
Genetic engineering wikipedia , lookup
Non-coding RNA wikipedia , lookup
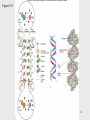
Molecular cloning wikipedia , lookup
Genome (book) wikipedia , lookup
Genomic library wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genome evolution wikipedia , lookup
Epitranscriptome wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of human development wikipedia , lookup
DNA supercoil wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome editing wikipedia , lookup
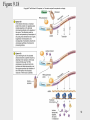
Genetic code wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding DNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
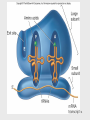
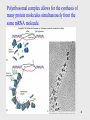
Helitron (biology) wikipedia , lookup
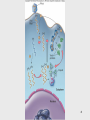
Nucleic acid analogue wikipedia , lookup
Microevolution wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Point mutation wikipedia , lookup
Primary transcript wikipedia , lookup
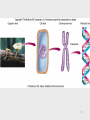
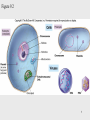
Lecture PowerPoint to accompany Foundations in Microbiology Seventh Edition Talaro Chapter 9 Microbial Genetics Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. 9.1 Genetics and Genes Genetics – the study of heredity The science of genetics explores: 1. Transmission of biological traits from parent to offspring 2. Expression and variation of those traits 3. Structure and function of genetic material 4. How this material changes 2 3 Levels of Structure and Function of the Genome • Genome – sum total of genetic material of a cell (chromosomes + mitochondria/chloroplasts and/or plasmids) – Genome of cells – DNA – Genome of viruses – DNA or RNA • DNA complexed with protein constitutes the genetic material as chromosomes • Bacterial chromosomes are a single circular loop • Eukaryotic chromosomes are multiple and linear 4 Figure 9.2 5 Chromosome is subdivided into genes, the fundamental unit of heredity responsible for a given trait – – Site on the chromosome that provides information for a certain cell function Segment of DNA that contains the necessary code to make a protein or RNA molecule Three basic categories of genes: 1. Genes that code for proteins – structural genes 2. Genes that code for RNA 3. Genes that control gene expression – regulatory genes 6 • All types of genes constitute the genetic makeup – genotype • The expression of the genotype creates observable traits – phenotype 7 Genomes Vary in Size • Smallest virus – 4-5 genes • E. coli – single chromosome containing 4,288 genes; 1 mm; 1,000X longer than cell • Human cell – 46 chromosomes containing 31,000 genes; 6 feet; 180,000X longer than cell 8 Figure 9.3 E. coli cell has spewed out its DNA 9 DNA • Two strands twisted into a double helix • Basic unit of DNA structure is a nucleotide • Each nucleotide consists of 3 parts: – A 5 carbon sugar – deoxyribose – A phosphate group – A nitrogenous base – adenine, guanine, thymine, cytosine • Nucleotides covalently bond to form a sugarphosphate linkage – the backbone – Each sugar attaches to two phosphates – • 5′ carbon and 3′ carbon 10 DNA • Nitrogenous bases covalently bond to the 1′ carbon of each sugar and span the center of the molecule to pair with an appropriate complementary base on the other strand – Adenine binds to thymine with 2 hydrogen bonds – Guanine binds to cytosine with 3 hydrogen bonds • Antiparallel strands 3′ to 5′ and 5′ to 3′ • Each strand provides a template for the exact copying of a new strand • Order of bases constitutes the DNA code 11 Figure 9.4 12 Significance of DNA Structure 1. Maintenance of code during reproduction - Constancy of base pairing guarantees that the code will be retained 2. Providing variety - order of bases responsible for unique qualities of each organism 13 DNA Replication • Making an exact duplicate of the DNA involves 30 different enzymes • Begins at an origin of replication • Helicase unwinds and unzips the DNA double helix • An RNA primer is synthesized at the origin of replication • DNA polymerase III adds nucleotides in a 5′ to 3′ direction – Leading strand – synthesized continuously in 5′ to 3′ direction – Lagging strand – synthesized 5′ to 3′ in short segments; overall direction is 3′ to 5′ 14 • DNA polymerase I removes the RNA primers and replaces them with DNA • When replication forks meet, ligases link the DNA fragments along the lagging strand to complete the synthesis • Separation of the daughter molecules is complete 15 Figure 9.5 16 DNA replication is semiconservative because each chromosome ends up with one new strand of DNA and one old strand. 18 Figure 9.7 Completion of chromosome replication 19 9.2 Applications of the DNA code • Information stored on the DNA molecule is conveyed to RNA molecules through the process of transcription • The information contained in the RNA molecule is then used to produce proteins in the process of translation 20 21 Gene-Protein Connection 1. Each triplet of nucleotides on the RNA specifies a particular amino acid 2. A protein’s primary structure determines its shape and function 3. Proteins determine phenotype. Living things are what their proteins make them. 4. DNA is mainly a blueprint that tells the cell which kinds of proteins to make and how to make them 22 Figure 9.9 DNA-protein relationship 23 RNAs • Single-stranded molecule made of nucleotides – 5 carbon sugar is ribose – 4 nitrogen bases – adenine, uracil, guanine, cytosine – Phosphate 24 RNA • 3 types of RNA: – Messenger RNA (mRNA) – carries DNA message through complementary copy; message is in triplets called codons – Transfer RNA (tRNA) – made from DNA; secondary structure creates loops; bottom loop exposes a triplet of nucleotides called anticodon which designates specificity and complements mRNA; carries specific amino acids to ribosomes – Ribosomal RNA (rRNA) – component of ribosomes where protein synthesis occurs 25 26 Figure 9.10 27 Transcription: The First Stage of Gene Expression 1. RNA polymerase binds to promoter region upstream of the gene 2. RNA polymerase adds nucleotides complementary to the template strand of a segment of DNA in the 5′ to 3′ direction 3. Uracil is placed as adenine’s complement 4. At termination, RNA polymerase recognizes signals and releases the transcript 100-1,200 bases long 28 Figure 9.11 29 Translation: The Second Stage of Gene Expression • All the elements needed to synthesize protein are brought together on the ribosomes • The process occurs in five stages: initiation, elongation, termination, and protein folding and processing 30 Figure 9.12 31 The Master Genetic Code • Represented by the mRNA codons and the amino acids they specify • Code is universal • Code is redundant 32 Figure 9.13 33 Figure 9.14 34 Translation • Ribosomes assemble on the 5′ end of an mRNA transcript • Ribosome scans the mRNA until it reaches the start codon, usually AUG • A tRNA molecule with the complementary anticodon and methionine amino acid enters the P site of the ribosome and binds to the mRNA 35 36 Translation • A second tRNA with the complementary anticodon fills the A site • A peptide bond is formed • The first tRNA is released and the ribosome slides down to the next codon • Another tRNA fills the A site and a peptide bond is formed • This process continues until a stop codon is encountered 37 Translation Termination • Termination codons – UAA, UAG, and UGA – are codons for which there is no corresponding tRNA • When this codon is reached, the ribosome falls off and the last tRNA is removed from the polypeptide 38 Figure 9.15 39 Polyribosomal complex allows for the synthesis of many protein molecules simultaneously from the same mRNA molecule. 40 Eukaryotic Transcription and Translation 1. Do not occur simultaneously – transcription occurs in the nucleus and translation occurs in the cytoplasm 2. Eukaryotic start codon is AUG, but it does not use formyl-methionine 3. Eukaryotic mRNA encodes a single protein, unlike bacterial mRNA which encodes many 4. Eukaryotic DNA contains introns – intervening sequences of noncoding DNA – which have to be spliced out of the final mRNA transcript 41 Figure 9.17 42 Genetics of Animal Viruses • Viral genome - one or more pieces of DNA or RNA; contains only genes needed for production of new viruses • Requires access to host cell’s genetics and metabolic machinery to instruct the host cell to synthesize new viral particles 43 44 45 9.3 Regulation of Protein Synthesis and Metabolism • Genes are regulated to be active only when their products are required • In prokaryotes this regulation is coordinated by operons, a set of genes, all of which are regulated as a single unit 46 Operons • 2 types of operons: – Inducible – operon is turned ON by substrate: catabolic operons - enzymes needed to metabolize a nutrient are produced when needed – Repressible – genes in a series are turned OFF by the product synthesized; anabolic operon – enzymes used to synthesize an amino acid stop being produced when they are not needed 47 Lactose Operon: Inducible Operon Made of 3 segments: 1. Regulator – gene that codes for repressor 2. Control locus – composed of promoter and operator 3. Structural locus – made of 3 genes each coding for an enzyme needed to catabolize lactose – b-galactosidase – hydrolyzes lactose permease – brings lactose across cell membrane b-galactosidase transacetylase – uncertain function 48 Lac Operon • Normally off – In the absence of lactose, the repressor binds with the operator locus and blocks transcription of downstream structural genes • Lactose turns the operon on – Binding of lactose to the repressor protein changes its shape and causes it to fall off the operator. RNA polymerase can bind to the promoter. Structural genes are transcribed. 49 Figure 9.18 50 Arginine Operon: Repressible • Normally on and will be turned off when the product of the pathway is no longer required • When excess arginine is present, it binds to the repressor and changes it. Then the repressor binds to the operator and blocks arginine synthesis. 51 Figure 9.19 52 9.4 Mutations: Changes in the Genetic Code • A change in phenotype due to a change in genotype (nitrogen base sequence of DNA) is called a mutation • A natural, nonmutated characteristic is known as a wild type (wild strain) • An organism that has a mutation is a mutant strain, showing variance in morphology, nutritional characteristics, genetic control mechanisms, resistance to chemicals, etc. 53 Figure 9.20 54 Causes of Mutations • Spontaneous mutations – random change in the DNA due to errors in replication that occur without known cause • Induced mutations – result from exposure to known mutagens, physical (primarily radiation) or chemical agents that interact with DNA in a disruptive manner 55 56 Categories of Mutations • Point mutation – addition, deletion, or substitution of a few bases • Missense mutation – causes change in a single amino acid • Nonsense mutation – changes a normal codon into a stop codon • Silent mutation – alters a base but does not change the amino acid 57 Categories of Mutations • Back-mutation – when a mutated gene reverses to its original base composition • Frameshift mutation – when the reading frame of the mRNA is altered 58 59 Repair of Mutations • Since mutations can be potentially fatal, the cell has several enzymatic repair mechanisms in place to find and repair damaged DNA – DNA polymerase – proofreads nucleotides during DNA replication – Mismatch repair – locates and repairs mismatched nitrogen bases that were not repaired by DNA polymerase – Light repair – for UV light damage – Excision repair – locates and repairs incorrect sequence by removing a segment of the DNA and then adding the correct nucleotides 60 Figure 9.21 61 The Ames Test • Any chemical capable of mutating bacterial DNA can similarly mutate mammalian DNA • Agricultural, industrial, and medicinal compounds are screened using the Ames test • Indicator organism is a mutant strain of Salmonella typhimurium that has lost the ability to synthesize histidine • This mutation is highly susceptible to backmutation 62 Figure 9.22 63 Positive and Negative Effects of Mutations • Mutations leading to nonfunctional proteins are harmful, possibly fatal • Organisms with mutations that are beneficial in their environment can readily adapt, survive, and reproduce – these mutations are the basis of change in populations • Any change that confers an advantage during selection pressure will be retained by the population 64 9.5 DNA Recombination Events Genetic recombination – occurs when an organism acquires and expresses genes that originated in another organism 3 means for genetic recombination in bacteria: 1. Conjugation 2. Transformation 3. Transduction 65 66 Conjugation • Conjugation – transfer of a plasmid or chromosomal fragment from a donor cell to a recipient cell via a direct connection – Gram-negative cell donor has a fertility plasmid (F plasmid, F′ factor) that allows the synthesis of a conjugative pilus – Recipient cell is a related species or genus without a fertility plasmid – Donor transfers fertility plasmid to recipient through pilus 67 Figure 9.23 (1) 68 Figure 9.23 (2) 69 Conjugation • High-frequency recombination – donor’s fertility plasmid has been integrated into the bacterial chromosome • When conjugation occurs, a portion of the chromosome and a portion of the fertility plasmid are transferred to the recipient 70 Figure 9.23 (3) 71 Transformation • Transformation – chromosome fragments from a lysed cell are accepted by a recipient cell; the genetic code of the DNA fragment is acquired by the recipient • Donor and recipient cells can be unrelated • Useful tool in recombinant DNA technology 72 Figure 9.24 Insert figure 9.23 transformation 73 Transduction • Transduction – bacteriophage serves as a carrier of DNA from a donor cell to a recipient cell • Two types: – Generalized transduction – random fragments of disintegrating host DNA are picked up by the phage during assembly; any gene can be transmitted this way – Specialized transduction – a highly specific part of the host genome is regularly incorporated into the virus 74 Figure 9.25 Generalized transduction 75 Figure 9.26 Specialized transduction 76 Transposons • Special DNA segments that have the capability of moving from one location in the genome to another – “jumping genes” • Cause rearrangement of the genetic material • Can move from one chromosome site to another, from a chromosome to a plasmid, or from a plasmid to a chromosome • May be beneficial or harmful 77 Figure 9.27 Transposons 78