* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download The Cell Cycle
Cell nucleus wikipedia , lookup
Cell membrane wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Endomembrane system wikipedia , lookup
Cell culture wikipedia , lookup
Extracellular matrix wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Programmed cell death wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell growth wikipedia , lookup
Cytokinesis wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Paracrine signalling wikipedia , lookup
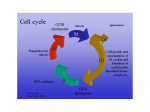
The Cell Cycle and its implications in diseases Hansjörg Hauser Dept. of Gene Regulation and Differentiation Molecular Biotechnology Helmholtz Centre for Infection Research, Braunschweig Cell division is a prerequisite for life •Microorganisms reproduce by cell division •Mammals need cell division during embryogenesis and for tissue homeostasis Example: Adult humans produce several milions of new cells per second (more than 1011 per day – about 100 grams) Cell division can be fast or slow •Microorganisms > 20 min per division •Multicellular organisms: 8 min and several weeks per division •All species can halt cell division The Cell Cycle M G2 G1 S S Start, G1 Checkpoint Point of no return Methods to measure cell division •Counting •Amount of DNA •Enzymatic activities •Incorporation of labeled DNA precursors •Cell cycle analysis (FACS) •Dilution of dyes •Time lapse microscopy While cdks are constitutively expressed the appearance of cyclins in the cell cycle is transient – they cycle The presence of cyclins regulates the activity of the cdks Cyclic activity of Cyclin kinases Temporal control of the animal cell cycle. The cyclin-E-, cyclin-A- and cyclin-B-dependent kinases are active at different times in the cell cycle. On this basis, cyclin E–Cdk2 appears to have a role in promoting S phase, cyclin A–Cdk2 in S phase and at G2-to-M phase, and cyclin B–Cdk1 during mitosis. The kinase activity of cdc-cyclin compexes is regulated by phosphorylation and dephosphorylation Examples MO16 is an activating kinase Wee1 is an inhibitory kinase cdc25 is a phosphatase that removes the inhibitory phosphate from the cdk Regulation of cyclin-dependent kinases. Arrowheads represent activating events and perpendicular ends represent inhibitory events. Genes known to perform the indicated functions are listed below. Both cyclins and some CKIs (Cdk inhibitors) are regulated by synthesis and ubiquitin-mediated proteolysis. Checkpoint pathways could act to promote inhibitory pathways or inhibit activating pathways to cause cell cycle arrest Example: The CKI (cdk inhibitor) p27 p27 inhibits cdk2 and thereby the relvant complexes CyclinE/cdk2 and CyclinA/cdk2 p27 underlies multiple regulations: transcriptional, translational, degradation, localisation (cytoplasmic versus nuclear), phosphorylation, sequestering by binding to CyclinD1/cdk4 (without becoming inactivated) The progression through the cell cycle underlies many controls: Example DNA replication A re-replication block ensures that no segment of DNA is replicated more than once Passage through mitosis removes the rereplication block Feedback controls generally depend on inhibitory signals Checkpoint pathways (A) A genetic pathway illustrating intrinsic and extrinsic checkpoint mechanisms. Letters represent cell cycle processes. The pathway shown as red symbols indicates an intrinsic checkpoint mechanism that operates to ensure that event C is completed before event E. After event B is completed, an inhibitory signal is activated that blocks completion of event E. After event C is completed, a signal is sent to turn off the inhibitory signal from B, thereby allowing completion of E. The blue symbols represent an extrinsic mechanism that is activated when defects such as DNA damage or spindle errors are detected. It is arbitrarily located on the D to E pathway but could also function by inhibiting a later step in the B to C pathway. In that case, the extrinsic pathway would utilize the intrinsic mechanism for cell cycle arrest. Mutations in any of the red or blue symbols would result in a checkpoint-effective phenotype. DNA damage leads to a block in cell cycle progression Replication of damaged DNA would fix mutations for all daughter cells Possible biochemical function of the Rad24 group of checkpoint proteins. Rad24, together with the four small subunits of RFC, is a component of a pentameric complex. By analogy with RFC, this complex might recognise the transition between ssDNA and dsDNA. Such a structure is produced by many repair pathways but the Rad24 complex may only efficiently recognise it in the context of repair complexes (not shown here). Once the Rad24 complex is bound, it then functions to recruit the ‘PCNA-like’ Rad17/Mec3/Ddc1 complex to the DNA, followed by additional recruitment of checkpoint proteins involved in signal transduction (e.g. Mec1 and Rad53) Gene expression in G1 G1 G0 Activation of early response genes: fos, jun,.. delayed response genes: E2F Cyclins E, D S DNA synthesis genes Resting cells: The retinoblastoma protein Rb blocks cell cycle progression in G1 by binding to and sequestering E2F Rb-P Rb + E2F Phosphorylation causes Inactivation of Rb Rb : E2F Target of the CyclinE/cdk2 and CyclinD/cdk4(6) complexes Phosphorylation of pRB Cell cycle progression by growth factors Phosphorylation causes Inactivation of Rb Rb-P Proliferation Rb E2F CyclinD.cdk4 CyclinD MAPK pathway Ras EGF Rb: E2F Rb captures E2F: E2F cannot activate proproliferative genes Proliferation Cell cycle progression Growth block Phosphorylation causes Inactivation of Rb Rb-P Rb captures E2F, so that it cannot activate proproliferative genes Rb CyclinD.cdk4 + E2F CyclinD Rb: E2F MAPK pathway Ras P16 Ink4A Regulators the cell cycle progression CDK1 CycB M CDK4/6 CycD pRB E2F P G2 INK4 p15 p16 p18 p19 P pRB G1 E2F CDK2 CycE CDK1 CycA S CDK2 CycA Kip/Cip p21 p27 p57 Mammalian cells: The protein p53 is sensing DNA damage p53 becomes phosphorylated and stabilized P53 is a general gatekeeper for the G1 checkpoint p53 is a transcriptional activator: One of the genes induced by p53 is p21, an inhibitor of the cdk4 kinase activity p27 Rb captures E2F, so that it cannot activate proproliferative genes CyclinE.cdk2 Rb-P p53 Rb CyclinD.cdk4 + E2F p21 CyclinD MAPK pathway Ras Rb: E2F p16 Ink4A Cellular products influencing the cell cycle Viral and cellular proteins influencing p53 activity Cell cycle control in mono- versus multicellular systems •Monocellular systems: Unlimited proliferation Control by size, nutrients and sex •Multicellular systems: Proliferation is limited to specific regions and circumstances: Growth factors, cell:cell-interactions, In mammals growth and proliferation are independently regulated Influence of cellular and viral proteins in the cell cycle machinery Growth factor stimulation through membrane receptors Extracellular ligand binding domain Transmembrane domain Tyrosine kinase domain EGF receptor FGF receptor Growth factor stimulation through a membrane receptor EGFreceptor Tyrosine kinase Cell growth inhibitors that act through a membrane receptor Anti-GrowthFactors e.g. TGFß p16 Cycl D:CDK4 RB E2Fs Changes in Gene Expression p15 Smads p27 Cycl E:CDK4 - Cell Proliferation (Cell Cycle) p21 Rb regulates the cell cycle by binding nuclear transcription factors E2F/myc Rb-P Rb + E2F Phosphorylation causes Inactivation of Rb TGFβ E2F Rb myc Cancer and the cell cycle Introduction Current view: • accumulation of multiple mutations within genes of a single cell • mutations confer a competitive advantage for cell growth and (de-) differentiation • mutations lead to initiation and progression of malignancies Proto-oncogenes • control cell proliferation and differentiation • are expressed in all subcellular compartments (nucleus, cytoplasm, cell surface) • act as protein kinases, growth factors, growth factor receptors, or membrane associated signal transducers Oncogenes • Mutations in proto-oncogenes alter the normal structure and/or expression pattern • Act in a dominant fashion gain of function Mechanisms of oncogene action Biochemically, there are three known mechanisms by which these genes act: • phosphorylation of proteins, with serine, threonine and tyrosine as substrates • signal transmission by GTPases • regulation of DNA transcription Tumor Suppressor Genes • Have normal, diverse functions to regulate cell growth in a negative fashion (restrain neoplastic growth; act as cellular “brakes”) • physical or functional loss of both alleles frees the cell from constraints imposed by their protein products loss of function What causes cancer? • Chemical Carcinogenes – Aflatoxin B1,, Vinylchloride, β-Propiolacton Dimethylsulfate ... • Radiation: – UV, X-Ray, α,-β,-γ-radiation • Viruses – RNA-viruses, DNA-viruses • Spontaneoud mutations Loss of DNA-repair machinery p53 „... manifestation of six essential alterations in cell physiology that collectively dictate malignant growth.“ Cell, Vol. 100, 2000 1. Self-Sufficiency in Growth Signals 2. Insensitivity to Antigrowth Signals 3. Evading Apoptosis 4. Limitless Replicative Potential 5. Sustained Angiogenesis 6. Tissue Invasion and Metastasis Summary „... manifestation of six essential alterations in cell physiology that collectively dictate malignant growth.“ Cell, Vol. 100, 2000 1. Self-Sufficiency in Growth Signals growth signals (PDGF, TGFα) --> autocrine stimulation overexpression or mutation of receptors (EGF-R, HER2) disruption of intracellular circuits (SOS-Ras-Raf -Map-Kinase) 2. Insensitivity to Antigrowth Signals Disruption of Rb-pathway, downregulation of death receptors 3. Evading Apoptosis Loss of proapoptotic regulators (p53), nonsignaling deathreceptors (FAS) 4. Limitless Replicative Potential Telomerase activation 5. Sustained Angiogenesis Increased expression of angiogen. inducers (VEGF, bFGF) loss of p53 -->downregulation of inhibitors (thrombospondin-1) 6. Tissue Invasion and Metastasis „Out-of-order“ CAMs (E-cadherin), changing integrin expression pattern, overexpression of extracellular proteases, downregulation of protease inhibitor genes