* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download File
SNARE (protein) wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Endomembrane system wikipedia , lookup
Protein moonlighting wikipedia , lookup
Signal transduction wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein structure prediction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
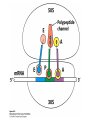
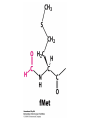
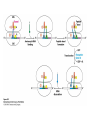
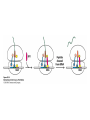
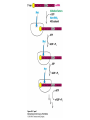
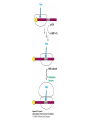
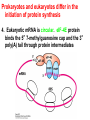
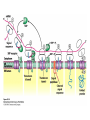
Protein synthesis decodes the information in messenger RNA Protein synthesis occurs in three phases: 1. Initiation – the translation machinery locates the start codon in mRNA 2. Elongation – codons are read 5’ 3’ as the protein is synthesized from the amino end to the carboxyl end 3. Termination – special proteins hydrolyze the polypeptide from the last tRNA when Ribosomes have three tRNA-binding sites that bridge the 30S and 50S subunits The mRNA being translated is bound to 30S Each tRNA molecule contacts both 30S and 50S Two of the three tRNAs have anticodon-codon interactions with the mRNA A site = aminoacyl site P site = peptidyl site The E site (exit site) contains the third tRNA tRNA acceptor stems are positioned in 50S The start signal is AUG or GUG preceded by several bases that pair with 16S rRNA Nearly half the amino terminal aa residues in proteins from E. coli are methionine, suggesting that AUG is the start codon Initiator regions contain a purine-rich sequence called the Shine-Dalgarno sequence about 10 nucleotides 5’ of the initiator codon The Shine-Dalgarno sequence interacts with a complementary region on the 3’ end of 16S RNA Bacterial protein synthesis is initiated by formylmethionyl (fmet) tRNA The initiator tRNA (tRNAf) differs from tRNAm used for internal methionine residues The initiator fmet is removed from about half of the proteins found in E. coli from the newly synthesized protein A single aminoacyl-tRNA synthetase links met to both tRNA molecules, however met attached to tRNAf is recognized by a specific enzyme that formylates the met amino group fMet-tRNAf is placed in the P site during formation of the 70S initiation complex Initiation factors IF1 and IF3 join the 30S subunit to preventing 30S from prematurely binding 50S IF2, a GTPase, binds GTP to change shape and enable binding of IF2 to fMet-tRNAf. The IF2-GTP- fMet-tRNAf complex binds mRNA bound to 16S rRNA via theShine-Dalgarno sequence to create the 30S initiation complex Binding of 50S causes GTP hydrolysis, IF’s are released and the 70S initiation complex is formed Elongation factors deliver aminoacyl-tRNA to the ribosome The mRNA codon in the A site defines which aminoacyl-tRNA will enter the site Elongation Factor Tu (EF-Tu) delivers the correct aminoacyl-tRNA to the A site when GTP is bound EF-Tu serves two functions: 1. EF-Tu protects the ester linkage in aminoacyltRNA from hydrolysis 2. The GTP in EF-Tu is hydrolyzed to GDP only when an appropriate complex between the EF-Tu-aminoacyl-tRNA complex and the ribosome has formed EF-Tu is then reset to its GTP form by EF-Ts which induces dissociation of GDP from EF-Tu and replacement by GTP Peptidyl transferase catalyzes peptide bond formation When both the A and P sites are occupied by aminoacyl-tRNA, the formylmethionine linked to initiator tRNA is transferred to the amino group in the A site. The peptidyl transferase center on the 23S subunit of the 50S subunit catalyzes formation of the peptide bond Formation of a peptide bond is followed by GTPdriven translocation of tRNAs and mRNA The translocation mechanism Proteins are synthesized by the successive addition of amino acids to the carboxyl terminus Protein synthesis is terminated by release factors that read stop codons Stop codons (UAA, UGA or UAG) are read by protein release factors FR1 recognizes UAA or UAG and RF2 recognizes UAA or UGA RF3 is a GTPase that mediates interactions between RF1 or RF2 and the ribosome RF1 and RF2 mimic tRNAs and promote hydrolytic attack on the ester linkage between tRNA and the polypeptide Prokaryotes and eukaryotes differ in the initiation of protein synthesis 1. Eukaryotic ribosomes are larger, consisting of a 60S large subunit and a 40S small subunit. The 60S subunit contains three RNAs: 5S RNA, 28S RNA and 5.8S RNA. The 40S subunit contains an 18S RNA. 2. The initiating amino acid in eukaryotes is methionine rather than N-formylmethionine. A special initiator tRNA is used called tRNAi 3. The initiating codon is always AUG with no Shine-Dalgarno sequence Prokaryotes and eukaryotes differ in the initiation of protein synthesis 4. Eukaryotic mRNA is circular. eIF-4E protein binds the 5’ 7-methylguanosine cap and the 3’ poly(A) tail through protein intermediates Some antibiotics inhibit proteins synthesis Streptomycin, a highly basic trisaccharide, interferes with binding of formylmethionyl-tRNA to ribosomes preventing initiation Neomycin, kanamycin and gentamycin interfere with interactions between tRNA and the 16S rRNA to inhibit initiation Choramphenicol inhibits peptidyl transferase Erythromycin binds the 50S subunit and blocks translocation Ribosomes bound to the endoplasmic reticulum manufacture secretory and membrane proteins Ribosomes attached to the endoplasmic reticulum to form the Rough Endoplasmic Reticulum (RER) Ribosomes on RER synthesize proteins destined to exit the cell or become membrane proteins exposed on the surface of the cell Three classes: Secretory proteins; lysosomal proteins; and proteins spanning the plasma membrane Protein synthesis begins on ribosomes that are free in the cytoplasm Secretory, lysosomal and membrane proteins begin synthesis on a free ribosome but then arrest until the ribosome binds the cytoplasmic surface of the ER Docking with the ER restarts protein synthesis with the newly synthesized protein threaded into the lumen of the ER Signal sequences mark proteins for translocation across the ER membrane The translocation machinery consists of four components: 1. The Signal Sequence – 9 to 12 hydrophobic amino acid residues, sometimes with positively charged amino acid residues, usually near the amino terminus of the nascent peptide chain. Some signal sequences are maintained in the mature protein while others are cleaved by a signal peptidase. 2. The Signal Recognition Particle (SRP) recognizes and binds the signal peptide then directs the peptide to the ER lumen. SRPs are GTPases. The translocation machinery consists of four components: 3. The SRP Receptor binds SRP at the surface of the ER membrane. The SRP Receptor is a GTPase. 4. The Translocon is the translocation machinery that transports the nascent polypeptide across the ER membrane.