* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 10 Analyzing the DNA by array and deep sequencing (1)
Extrachromosomal DNA wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Epigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Population genetics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Neocentromere wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Behavioural genetics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Copy-number variation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Human genetic variation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
Human genome wikipedia , lookup
Metagenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Oncogenomics wikipedia , lookup
Medical genetics wikipedia , lookup
Genome (book) wikipedia , lookup
Human Genome Project wikipedia , lookup
Genomic library wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Public health genomics wikipedia , lookup
Genome editing wikipedia , lookup
Microevolution wikipedia , lookup
Exome sequencing wikipedia , lookup
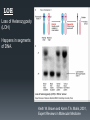
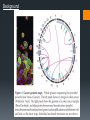
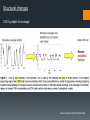
Analyzing DNA using Microarray and Next Generation Sequencing (1) Background SNP Array Basic design Applications: CNV, LOH, GWAS Deep sequencing Alignment and Assembly Applications: structural changes, GWAS The chromosome SNP Variations in DNA sequence. Single Nucleotide Polymorphism (SNP) --- a single letter change in the DNA. Common SNPs occur every few hundred bases. Each form is called an “allele”. Almost all SNPs have only two alleles. Allele frequencies are often different between ethnic groups. http://upload.wikimedia.org/wiki pedia/commons/thumb/2/2e/Dn a-SNP.svg/180px-DnaSNP.svg.png Correlations between SNPs Why measure the SNP alleles? DNA change in two ways during evolution: Point mutation SNPs Recombination This happens in large segments. Alleles of adjacent SNPs are highly dependent. Haplotype: A group of alleles linked closely enough to be inherited mostly as a unit. http://www.evolutionpages.com/images/ crossing_over.gif Why SNP? http://www.hapmap.org/originhaplotype. html.en Figure 1: This diagram shows two ancestral chromosomes being scrambled through recombination over many generations to yield different descendant chromosomes. If a genetic variant marked by the A on the ancestral chromosome increases the risk of a particular disease, the two individuals in the current generation who inherit that part of the ancestral chromosome will be at increased risk. Adjacent to the variant marked by the A are many SNPs that can be used to identify the location of the variant. Why SNP? Nature Genetics 26, 151 - 157 (2000) SNPs Figure 1. Schematic model of trait aetiology. The phenotype under study, Ph, is influenced by diverse genetic, environmental and cultural factors (with interactions indicated in simplified form). Genetic factors may include many loci of small or large effect, GPi, and polygenic background. Marker genotypes, Gx, are near to (and hopefully correlated with) genetic factor, Gp, that affects the phenotype. Genetic epidemiology tries to correlate Gx with Ph to localize Gp. Above the diagram, the horizontal lines represent different copies of a chromosome; vertical hash marks show marker loci in and around the gene, Gp, affecting the trait. The red Pi are the chromosomal locations of aetiologically relevant variants, relative to Ph. The gene deciding pheonotype SNP array The SNP array Affymetrix.com SNP array The SNP array 40 probes per SNP (20 for forward strand and 20 for reverse strand.) PM/MM strategy. Data summary (generating AA/AB/BB calls) omitted here. Affymetrix.com SNP array Association analysis Genotype calls Linkage analysis SNP array Loss of Heterozygosity Signal strength Copy number abberation CNA --- Background Copy Number Aberration (CNA): A form of chromosomal aberration Deviation from the regular 2 copies for some segments of the chromosomes One of the key characteristics of cancer CNA in cancer: Reduce the copy number of tumor-suppressor genes Increase the copy number of oncogenes Possibly related to metastasis CNA --- the statistician’s task High density arrays allow us to identify “focused CNA”: copy number change in small DNA segments. With the high per-probeset noise, how to achieve high sensitivity AND specificity? CNA – maximizing sensitivity/specificity Two approaches that complement each other: Reducing noise at the single probeset level: Based on dose-response (Huang et al., 2006) Based on sequence properties (Nannya et al., 2005) Segmentation methods. Smoothing; Hidden Markov Model-based methods; Circular Binary Segmentation … … HMM data segmentation Fridlyand et al. Journal of Multivariate Analysis, June 2004, V. 90, pp. 132-153 Amplified Normal Deleted Forward-backword fragment assembling Some example: Top: model cell line, 3 copy segment in chromosome 9 Bottom: Cancer sample LOH Loss of Heterozygosity (LOH) Happens in segments of DNA. Keith W. Brown and Karim T.A. Malik, 2001, Expert Reviews in Molecular Medicine LOH On SNP array, LOH will yield identical calls (AA or BB, rather than AB) for a number of consecutive SNPs. Discov Med. 2011 Jul;12(62):25-32. GWAS http://www.mpg.de/10680/Modern_psychiatry © Pasieka, Science Photo Library GWAS GWAS Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer Nature Genetics 41, 986 - 990 (2009) DNA sequencing Background Background Background Background Alignment and Assembly When a reference genome is available --- Alignment Can rely on existing reference genome as a blue print. Align the short reads onto the reference genome. Need a few fold coverage to cover most regions. Sequence a whole new genome? --- Assembly Overlaps are required to construct the genome. The reads are short need ~30 fold coverage. If 3G data per run, need 30 runs for a new genome similar to human size. Alignment and Assembly Hash table-based alignment. Similar to BLAST in principle. (1) Find potential locations: (2) Local alignment. Alignment and Assembly From read to graph: Alignment and Assembly Alignment and Assembly de Bruijn graph assembly Red: read error. Alignment and Assembly de Bruijn graph assembly Alignment and Assembly de Bruijn graph assembly Whole gnome/exome/transcriptome sequencing Genomics Whole genome sequencing detects all variants (SNP alleles, rare variants, mutations) Could be associated with disease: Rare variants (burden testing by collapsing by gene) De novo mutations (need family tree) Rare Mendelian disorders Structural variants in cancer Structural changes Identification of translocations from discordant paired-end reads. Cancer Genetics 206 (2014) 432e440 Structural changes CNV by depth of coverage Cancer Genetics 206 (2014) 432e440 Structural changes Cancer Genetics 206 (2014) 432e440 Genotype calling http://www.geneious.com/features/sequence-analysis-annotation-prediction Medical Genomics Example: Extreme-case sequencing to find rare variants associated with a disease. Nature Reviews Genetics 11, 415 GWAS GWAS