* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A-Ag
Silencer (genetics) wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
Capillary electrophoresis wikipedia , lookup
Protein (nutrient) wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Signal transduction wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Biochemistry wikipedia , lookup
Circular dichroism wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Community fingerprinting wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Interactome wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein purification wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Western blot wikipedia , lookup
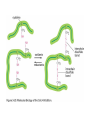
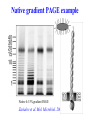
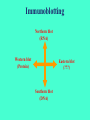
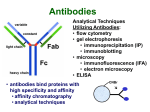
ELECTROPHORESIS 1. 2. 3. 4. 5. 6. Definitions Theory of Electrophoresis Electrophoretic Technique General Procedures Types of Electrophoresis Technical Considerations Agarose Gel Electrophoresis Gel electrophoresis is a widely used technique for the analysis of nucleic acids and proteins. Agarose gel electrophoresis is routinely used for the preparation and analysis of DNA. Gel electrophoresis is a procedure that separates molecules on the basis of their rate of movement through a gel under the influence of an electrical field. We will be using agarose gel electrophoresis to determine the presence and size of PCR products. 1. DEFINITIONS Electrophoresis – Migration of charged solutes in a liquid medium under an electrical field – Many biological molecules have ionisable groups eg. amino acids, proteins, nucleotides, nucleic acids – Under an electric field -> charged particles migrate to anode (+) or cathode (-) Zone Electrophoresis • Migration of charged molecules • Support medium – porous eg. CA or agarose – can be dried & kept • Same pH & field strength thru’ought • Separation based on electrophoretic mobility • Separates macromolecular colloids eg. proteins in serum, urine, CSF, erythrocytes; nucleic acids Isotachophoresis • Migration of small ions • Discontinuous electrolyte system – leading electrolyte (L- ions, high mobility) & – trailing electrolyte (T- ions) • Apply sample solution at interphase of L & T • Apply electric field -> each type of ion arrange between L and T ions -> discrete zones • Separates small anions, cations, organic & amino acids, peptides, nucleotides, nucleosides, proteins • Rate of migration depends on: – Net electrical charge of molecule – Size & shape of molecule – Electric field strength – Properties of supporting medium – Temperature of operation 3. ELECTROPHORETIC TECHNIQUE 3a. Instrumentation & Reagents • Buffer boxes with buffer plates -> holds buffer • Platinum or carbon electrode -> connected to power supply • Electrophoresis support -> with wicks to contact buffer • Cover -> minimize evaporation (Fig 7-1) 3c. Buffers • To carry applied current & to fix the pH => determine electrical charge & extent of ionization => which electrode to migrate • Ionic strength of buffer – thickness of ionic cloud -> migration rate -> sharpness of electrophoretic zones – [ion] -> ionic cloud -> movement of molecules • Barbital buffers & Tris-boric acid-EDTA buffers 3d. Protein Stains • To visualize/locate separated protein fractions • Dyes: amount taken up depends on – Type of protein – Degree of denaturation of proteins by fixing agents Types of stains: Table 7-1 4. GENERAL PROCEDURES 4a. Separation • Place support material in EP chamber • Blot excess buffer from support material • Place support in contact with buffer in electrode chamber • Apply sample to support cont. Separation • Separate component using constant voltage or constant current for length of time • Remove support, then -> dry or place in fixative -> treat with dye-fixative -> wash excess dye -> dry (agarose) or put in clearing agent (CA membs) 4b. Detection & Quantitation • Express as – % of each fraction present or – absolute concn • By densitometry – electrophoretic strip moved past an optical system – absorbance of each fraction displayed on recorder chart 5. TYPES OF ELECTROPHORESIS a. Agarose Gel Electrophoresis b. Cellulose Acetate Electrophoresis c. Polyacrylamide Gel Electrophoresis d. Isoelectric Focusing e. Two-dimensional Electrophoresis 5a. Agarose Gel Electrophoresis (AGE) • Use agarose as medium – low concns -> large pore size – higher concns -> small pore size • Serum proteins, Hb variants, lactate dehydrogenase, CK isoenzymes, LP fractions • Pure agarose - does not have ionizable groups -> no endosmosis Cont. AGE • Advantages: – low affinity for proteins – shows clear fractions after drying – low melting temp -> reliquify at 65oC • Disadvantage: – poor elasticity -> not for gel rod system -> horizontal slab gels 5b. Cellulose Acetate Electrophoresis (CAE) • Cellulose + acetic anhydride -> CA • Has 80% air space -> fill with liquid when soaked in buffer • Can be made transparent for densitometry • Advantages: – speed of separation – able to store transparent membranes • Disadvantages: – presoaking before use – clearing for densitometry cont. CAE • Method: – wet CA in EP buffer – load sample about 1/3 way along strip – stretch CA in strips across a bridge – place bridge in EP chamber -> strips dip directly into buffer – after EP, stain, destain, visualise proteins • For diagnosis of diseases – change in serum protein profile 5c. Polyacrylamide Gel Electrophoresis (PAGE) • Tubular-shaped EP cell -> pour small-pore separation gel -> large-pore spacer gel cast on top -> large-pore monomer solution + ~3ul sample on top of spacer gel • Electrophoresis -> all protein ions migrate thru large-pore gels -> concentrate on separation gel -> separation due to retardation of some proteins • Average pore size in 7.7% PAGE separation gel about 5nm -> allow most serum proteins to migrate -> impedes migration of large proteins eg fibrinogen, 1-lipoprotein, 2macroglobulin • Advantages: – thermostable, transparent, strong, chemically inert – wide range of pore sizes – uncharged -> no endosmosis • Disadvantages: – carcinogenic Denaturing PAGE/SDS-PAGE What is SDS-PAGE? • Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis • A procedure to separate proteins and determine their Molecular Weights. What is so special about SDS? • SDS is a negatively charged detergent. • Disrupts secondary and tertiary protein structures by breaking hydrogen bonds and unfolding protein. • ‘Masks’ charge on protein so that all proteins act the same as regards charge. • Prevents protein aggregation. • Prevents protein shape from influencing gel run. (i) Denaturing PAGE/SDS-PAGE Boil sample for 5 mins in sample buffer containing -mercaptoethanol & SDS -mercaptoethanol: reduce disulfide bridges SDS: binds strongly to & denatures proteins Proteins denatured -> opens into rodshaped structures -> separate based on size Use: – To assess purity of protein – To determine MW of protein (ii) Native PAGE • Use non-denaturing conditions -> no SDS or -mercaptoethanol -> proteins not denatured • Proteins separate based on: – different electrophoretic mobilities – sieving effects of gel • Use – to obtain native protein/enzyme – to study biological activity Native gradient PAGE example Native 4-15% gradient PAGE Zavialov et al. Mol. Microbiol. 2002 5d. Isoelectric Focusing • To separate amphoteric cpds eg. proteins • Proteins moves to zone where: pH medium = pI protein => charge = 0 • pI of protein confined in narrow pH range -> sharp protein zones • Method: – use horizontal gels on glass/plastic sheets – introduce ampholytes into gel -> create pH gradient cont. IEF Method – – – – – apply a potential difference across gel anode -> area with lowest pH cathode -> area with highest pH proteins migrate until it arrives at pH = pI wash with fixing solution to remove ampholytes – stain, destain, visualise • Separations of proteins with 0.01 to 0.02pH unit differences (Fig 7-4) 5e. Two-Dimensional (2D) EP (ISO-DALT) • 1st D using IEF EP -> in large-pore medium -> ampholytes to yield pH gradient • 2nd D using molecular weight-dependent EP -> in polyacrylamide -> linear or gradient • O’Farrell method: – use -mercaptoethanol (1st D) & SDS (2nd D) • Detect proteins using Coomassie dyes, silver stain, radiography, fluorography • Separates 1100 spots (autoradiography) Medium pH range (pH 4-7) pI 4 kDa 116 97 81 66 55 45 30 21 14 5 6 7 Narrow pH range (1 pH unit) (4.5-5.5) (4.0-5.0) (5.0-6.0) pI 4.0 116 97 81 66 55 45 MW (kDa) 30 21 14 4.5 5.0 5.5 6.0 SDS-PAGE buffers and Solutions Resolving buffer: Stacking buffer: 10% APS: 10X SDS-PAGE Tris-Glycine-SDS running buffer: 3X SDS-PAGE loading buffer: 30% 37.5:1 acrylamide/bisacrylamide solution: Immunoblotting Northern blot (RNA) Western blot (Protein) Eastern blot (???) Southern blot (DNA) IMMUNOASSAYS 1. Basic Concepts & Definitions 2. Measurement of Antibody Affinity 3. Quantitative Methods – competitive & noncompetitive assays 1. BASIC CONCEPTS & DEFINITIONS Immunoassay: use of antibodies to detect analyte 1a. Antibodies • Immunoglobulins that bind to Antigens • 5 classes: IgG, IgA, IgM, IgD, IgE 1b. Immunogen • Protein or a substance coupled to a carrier • When introduced into foreign host -> induce Ab to form 1c. Antigen • Any material which can react with Ab • May not induce Ab formation 1d. Antigen-Antibody Binding • Ab molecules have specific binding sites -> bind tightly to Ag -> cause pptn/neutralization/ death • Binding of Ag to Ab due to – van der Waals forces – hydrophobic interactions – charged group attractions • Can measure Antibody affinity: strength of binding between Ab & Ag 2. MEASUREMENT OF ANTIBODY AFFINITY • Binding of Ag to Ab is reversible -> association & dissociation Ag + Ab <-> AgAb • Law of mass action: Rate of rxn to concn of reactants ka[Ag][Ab] = kd[AgAb] K = ka/kd = [AgAb]/ [Ag][Ab] where K is equilibrium constant or affinity constant r/c = nK – rK r = no. of molecules of bound Ag per Ab molecule c = concn of free Ag n = valency of Ab • Plot r/c vs r => Scatchard Plot – Straight line with slope k – x intercept gives n – y intercept gives nK • K (liters/mole) measures affinity of complex Why measure Affinity of an Antibody? • To assess Ab specificity • It influences the functional efficiencies of Abs eg. high-affinity Abs are very dependable for various applications: – Diagnostic – Therapeutic – Analytical 3. QUANTITATIVE METHODS • Read & Understand from Tietz Fundamentals: – Radial Immunidiffusion Immunoassay – Electroimmunoassay – Turbidimetric & Nephelometric Assays • Labeled Immunochemical Assays COMPETITIVE vs NONCOMPETITIVE RXNS A. Competitive Immunoassays • Used when have limited reagents (Ag) (i) Simultaneous Competitive Assay • Labels Ag (Ag*) & unlabeled Ag compete for binding to Ab • The probability of Ab binding to Ag* is inversely to [Ag] Ab + Ag + Ag* <-> Ab:Ag + A-Ag* (ii) Sequential Competitive Assay • Step 1: unlabeled Ag mixed with excess Ab -> binding allowed to reach equilibrium • Step 2: Ag* added sequentially -> equilibrate • After separation -> det bound Ag* -> calculate [Ag] • Larger fraction of Ag bound to Ab than in simultaneous assay • If k1 >> k2 -> in Ab:Ag -> in Ag* binding • Provide two- to four- fold improvement in detection limit b. Noncompetitive Immunoassays • i. Used when have excess reagent Immobilization of Ab to support – Passively adsorption or bind covalently – Direct or indirect attachment (Table 9-3) ii. Ag allowed to react with Ab -> wash other proteins iii. Add labeled Ab (conjugate) -> reacts with bound Ag iv. Determine bound label -> [Ag*] or its activity is [Ag]