* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 2960 Lab 2 NOTES
Signal transduction wikipedia , lookup
Endomembrane system wikipedia , lookup
Protein moonlighting wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
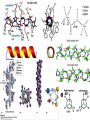
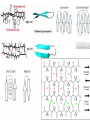
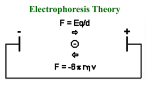
+ Lab 2: Organelles, Proteins, and SDS-PAGE Dr. Sarah VanVickle-Chavez BIO 2960 Laboratory Spring 2013 + Overview Today we will load a gel and compare your gel to the sample gel. Each person will load one lane on the gels in the center of each bench. While gels are running, we will do the PyMOL computer lab as a group demo. You should have Lab Report 1a complete before we leave today. You can use your books and notes, but there is a brief computer tutorial on the NSLC website to help you with Lab Report 1b. http://www.nslc.wustl.edu/courses/Bio2960/labs.html Next week, there are three reports due at the beginning of class 1a = proteins (PyMOL demo in class) 1b = cell organelles (use your book/notes for this one) 2 = SDS-PAGE + 1. Protein Electrophoresis SDS-PAGE stands for sodium dodecyl sulfate polyacrylamide gel electrophoresis + SDS vs Native Gel How do you “see” your proteins? + PyMOL Remember: Amino acids have different properties based on the R-group GAVLIMPFW = nonpolar STNQYC = uncharged, polar KRHDE = charged, polar + Alpha helix + Beta Sheet