* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bi-150-molbiol
Lipid signaling wikipedia , lookup
Biochemical cascade wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetic code wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Metalloprotein wikipedia , lookup
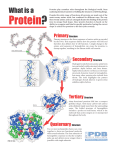
Magnesium transporter wikipedia , lookup
Gene regulatory network wikipedia , lookup
Epitranscriptome wikipedia , lookup
Interactome wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Point mutation wikipedia , lookup
Paracrine signalling wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Signal transduction wikipedia , lookup
Western blot wikipedia , lookup
Expression vector wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
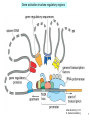
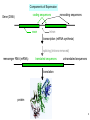
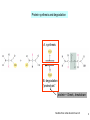
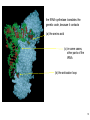
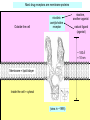
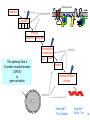
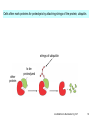
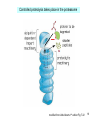
Bi 150 Lecture 0 October 4, 2012 An introduction to molecular biology . . . but you will learn the cell biology in this course 1 3 x 109 base pairs Lander et al Humans have 22 pairs of chromosomes, plus the X and Y. Males are XY; females are XX. A. Each chromosome is “painted” with a unique combination of fluorescent dyes B. We have arranged the chromosomes to form pairs. © Garland; Little Alberts Fig 5-12 Genes can be localized crudely by hybridizing a fluorescent nucleotide probe to chromosomes 2 mm 6 distinct genes are probed in this image Little Alberts Fig 10-16 Seuss 1959 Complete DNA sequence as scripture (surf NCBI) single-nucleotide polymorphisms (SNPs) chromosomal location orthologs in other species mutations that cause disease basic sequence RNA splicing RNA sequence proteins that bind to the sequence and regulate expression RNA abundance protein sequence protein function protein structure 5 How much coding sequence is in the genome? 22,000 genes x 400 codons/protein x 3 bases/codon = 26.4 million base pairs, or < 1% of the genome! The remainder . . . 1. Repetitive elements (junk? selfish DNA?) 2. Regulatory regions 3. Introns 6 Gene activation involves regulatory regions Little Alberts Fig. 8-15 © Garland publishing 7 Components of Expression coding sequences Gene (DNA) exon noncoding sequences intron transcription (mRNA synthesis) splicing (introns removed) messenger RNA (mRNA) translated sequences untranslated sequences translation protein 8 Protein synthesis and degradation A. synthesis B. degradation “proteolysis” protein + Greek, breakdown Modified from Little Alberts Panel 2-5 9 the tRNA synthetase translates the genetic code, because it contacts (a) the amino acid (c) in some cases, other parts of the tRNA (b) the anticodon loop 10 receptor a molecule on the cell surface or in the cell interior that has an affinity for a specific molecule (the ligand). Latin, “to tie” Most drug receptors are proteins. Greek, “first” 11 shortest: 9 longest: 5500 “peptide” or amide bonds 20 types side chains link the “backbone” or “main chain” or “a-carbons” Little Alberts Figure 2-22 © Garland publishing 12 Proteins contain a few structural motifs: a helices b sheets (A protein viewer must be installed on your computer) http://www.its.caltech.edu/~lester/Bi -1/alpha-helix-alphabetical.pdb http://www.its.caltech.edu/~lester/Bi -1/beta-sheet-antiparallel.pdb Hide side chains Show H-bonds and distances Show ribbons & arrows Show side chains Show Van der Waals radii 13 Most drug receptors are membrane proteins Outside the cell nicotinic acetylcholine receptor nicotine, another agonist natural ligand (agonist) ~ 100 Å = 10 nm Membrane = lipid bilayer Inside the cell = cytosol (view in ~1995) 14 Protein Folding vs. “Inverse Folding” = Computational Protein Design Protein Folding (no degeneracy) Set of All Structures Inverse Folding (large degeneracy) Individual amino acids Set of All Sequences Several ways to make an arch 15 A future Lecture outside receptor membrane b G protein i q s t a b a inside effector channel enzyme The pathway from a G protein-coupled receptor (GPCR) to gene activation intracellular messenger Ca2+ cAMP cytosol kinase phosphorylated protein nucleus How fast? 10 s to days How far? Up to 1 m 16 Protein degradation is accomplished primarily by proteolytic enzymes The genome encodes hundreds of proteolytic enzymes. They vary in -- sequence specificity for the “cut” -- cellular expression -- organelle of expression 17 Cells often mark proteins for proteolysis by attaching strings of the protein, ubiquitin. strings of ubiquitiin other protein to be proteolyzed modified from Little Alberts Fig 18-7 18 Controlled proteolysis takes place in the proteasome shorter modified from Little Alberts 1st edition Fig 7-32 19 Atomic-scale Structures H3CH 2C N morphine procaine nicotine CH 3 N N CH 2CH 3 H2C botulinum toxin HO O CH 2 N CH 3 O C O HO morphine NH 2 (Download to your computer; Then open with Swiss-prot pdb viewer) http://www.its.caltech.edu/~lester/Bi-1/morphine.pdb http://www.its.caltech.edu/~lester/Bi-1/procaine.pdb http://www.its.caltech.edu/~lester/Bi-1/nicotine.pdb 20