* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Cell cycle: Checkpoint proteins and kinetochores
Survey
Document related concepts
Cell nucleus wikipedia , lookup
Endomembrane system wikipedia , lookup
Tissue engineering wikipedia , lookup
Extracellular matrix wikipedia , lookup
Microtubule wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Signal transduction wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cell culture wikipedia , lookup
Cell growth wikipedia , lookup
List of types of proteins wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Cytokinesis wikipedia , lookup
Transcript
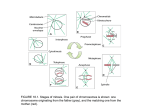
Dispatch R613 Cell cycle: Checkpoint proteins and kinetochores Aaron F. Straight Vertebrate homologs of yeast spindle assembly checkpoint proteins are localized to kinetochores and may act as a sensor for proper chromosome attachment to the mitotic spindle. Address: Department of Physiology, Box 0444, School of Medicine, University of California, San Francisco, California 94143-0444, USA. E-mail: [email protected] Current Biology 1997, 7:R613–R616 http://biomednet.com/elecref/09609822007R0613 © Current Biology Ltd ISSN 0960-9822 Cells carefully monitor the alignment of chromosomes on the mitotic spindle to ensure that their chromosomes are properly segregated at anaphase. Detachment of kinetochores from microtubules or depolymerization of the mitotic spindle activates a checkpoint mechanism that arrests the cell cycle prior to the initiation of anaphase. This ‘spindle assembly checkpoint’ allows the cell time to correct the offending lesion so that it can proceed through mitosis without chromosome missegregation [1,2]. Proteins involved in the spindle assembly checkpoint have been isolated in budding yeast and in vertebrates [3–7]. Cells lacking components of the spindle assembly checkpoint can proceed through mitosis in the absence of a spindle or in the presence of unattached chromosomes [3,5,8]. Recent work has shown that two of the spindle assembly checkpoint proteins, Mad2 and Bub1, localize to the kinetochores of vertebrate cells for part of mitosis. Prophase and prometaphase chromosomes show kinetochore localization of Mad2 and Bub1, but as chromosomes align on the metaphase plate the kinetochore staining disappears and does not reappear until the next cell cycle (Figure 1a). Misaligned chromosomes show Mad2 and Bub1 localization at the kinetochore even when all other chromosomes reside on the metaphase plate (Figure 1b,c) [4,6,7]. These results suggest that the kinetochore both monitors the attachment of the chromosome to the spindle and signals the cell to arrest until chromosomes are properly aligned. The spindle assembly checkpoint Early experiments in the 1930s showed that, when vertebrate cells are treated with drugs that depolymerize the mitotic spindle, they arrest in mitosis [9,10]. The mitotic arrest in these cells was attributed to the removal of the mechanical apparatus that separates the chromosomes — the mitotic spindle. Recent work has shown that the absence of the spindle itself is not the cause of the mitotic arrest, but that the arrest is mediated through the activation of a checkpoint that monitors the integrity of the spindle and the alignment of the chromosomes. Cells with an intact mitotic spindle will still arrest in mitosis if a single chromosome is not properly attached to the spindle (Figure 1b) [11]. This arrest is caused by an inhibitory signal emanating from kinetochores that are not attached to the spindle apparatus [12]. Genetic studies in yeast have identified some of the molecules involved in the spindle assembly checkpoint. Like vertebrate cells, budding yeast cells arrest in mitosis in response to spindle depolymerization [13]. Yeast mutants were isolated that died rapidly when treated with the spindle-depolymerizing drug benomyl. These mutants — mad1, mad2 and mad3, and bub1, bub2 and bub3 — all fail to activate the spindle assembly checkpoint and continue through mitosis without a spindle. Although the mutants die rapidly in the presence of benomyl, the mad and bub gene products do not appear to be directly involved in spindle integrity, as the mutant strains have normal spindles [3,5]. Instead, mad and bub mutants are thought to be defective in the signalling mechanism that arrests the cell cycle in response to spindle depolymerization. Wells and Murray [8] showed that excess kinetochores in budding yeast cells cause pronounced cell cycle delays in mitosis. These cell cycle delays are absent in the mad and bub mutants, suggesting that the Mad and Bub proteins are involved in monitoring chromosome attachment to kinetochores as well as spindle integrity. The kinetochore as the mechanism and the sensor for mitotic spindle attachment Kinetochores mediate attachment of the chromosomes to the mitotic spindle. When a microtubule contacts a kinetochore, the kinetochore captures the microtubule [14,15]. These kinetochore–microtubule attachments rearrange until sister kinetochores are attached to opposite poles of the spindle. In insect cells, tension on the kinetochores further stabilizes kinetochore–microtubule interactions, thus promoting bipolar attachment of the chromosomes to the spindle [16,17]. Once kinetochores are attached to the spindle microtubules, the kinetochores mediate congression of the chromosomes to the metaphase plate [18] and cells do not initiate anaphase until all chromosomes are properly aligned. Kinetochores can sense proper attachment to the mitotic spindle. Proteins at the kinetochore are specifically phosphorylated dependent upon their proper attachment to the mitotic spindle. In insect cells, kinetochores that are not attached to the mitotic spindle, and kinetochores that have only attached to one pole of the spindle, stain R614 Current Biology, Vol 7 No 10 Figure 1 (a) Interphase Prophase Mad Bub Metaphase Anaphase Misaligned chromosome Error correction Mad Bub M Buad b M Buad b (b) Mad Bub Mad Bub M Buad b Mad Bub M Buad b (c) Mitotic arrest Spindle depolymerization Mad Bub M Buad b M Buad b Mad Bub Mad Bub M Buad b Adaptation Mad Bub M Buad b Mitotic arrest © 1997 Current Biology Mad2 and Bub1 proteins reside at kinetochores and mediate checkpoint arrest of the cell cycle. (a) During a normal mitosis, Mad2 and Bub1 localize to kinetochores in prophase and prometaphase. Once all chromosomes are attached to the spindle, cells proceed through anaphase. (b) Misaligned chromosomes signal spindle assembly checkpoint arrest. Chromosomes that are not properly attached to the spindle maintain Mad2 at the kinetochore and the cell arrests in mitosis. (c) Spindle depolymerization causes mitotic arrest. In the absence of a spindle, Mad2 and Bub1 remain at the kinetochores of chromosomes and the cell arrests in mitosis. The checkpoint arrest can be overcome by two mechanisms: by error correction, the cell can correct the offending lesion and process through a normal mitosis; by adaptation, the cell can override the checkpoint arrest, in the presence of the defect, resulting in an abnormal mitosis with the potential for chromosome loss. brightly with the 3F3/2 antibody, which recognizes a phospho-epitope at the kinetochores of mammalian and insect cells. When chromosomes achieve a bipolar attachment to the spindle and the sister kinetochores come under tension, the staining of the 3F3/2 epitope is diminished. This loss of 3F3/2 staining can be mimicked by pulling on a mono-attached chromosome so as artificially to apply tension to the kinetochore. Cells that are microinjected with the 3F3 antibody delay dephosphorylation of the kinetochore and pause in mitosis. Thus, the kinetochore responds to proper attachment to the spindle by modifying the resident kinetochore proteins (reviewed in [19]). The spindle assembly checkpoint meets the kinetochore Three recent papers report that spindle assembly checkpoint proteins reside at the kinetochore and regulate passage through mitosis. Chen et al. [6] and Li and Benezra [4] isolated, respectively, the Xenopus (XMAD2) and human (hsMAD2) homologs of the yeast Mad2, and Taylor and McKeon [7] isolated the mouse homolog (mBub1) of yeast Bub1. All of these proteins localize to Dispatch the kinetochores of chromosomes in cultured cells of the organisms from which they were isolated. Interestingly, in all three organisms, the checkpoint proteins only localize to the kinetochores of cells in prophase and prometaphase, prior to proper chromosome alignment on the metaphase plate, or of cells that have been treated with nocodazole to depolymerize the mitotic spindle (Figure 1a,c). To investigate further the relationship between chromosome attachment to the spindle and XMAD2 staining, Chen et al. [6] took advantage of cells that had attached all but one or two chromosomes to both spindle poles. Chromosomes that had only attached a single kinetochore to the spindle showed no XMAD2 staining on the attached kinetochore, but bright XMAD2 staining on the unattached sister kinetochore (Figure 1b). Thus, the checkpoint proteins at the kinetochore respond to microtubule attachment to the kinetochore. Mad2 and Bub1 are required for the spindle assembly checkpoint in vertebrate cells. Chen et al. [6] and Li and Benezra [4] showed that antibodies against Mad2 blocked the spindle assembly checkpoint. The spindle assembly checkpoint can be reconstituted in vitro using extracts of Xenopus laevis eggs [20]. Using X. laevis egg extracts, Chen et al. [6] showed that the addition of anti-XMAD2 antibodies to a checkpoint-arrested extract inactivated the checkpoint and allowed progression through mitosis. Additionally, cells that were treated with anti-XMAD2 prior to checkpoint activation were unable to establish a checkpoint arrest. Li and Benezra [4] took a related approach to test the function of hsMAD2 in tissue culture cells. Antibodies to hsMAD2 were electroporated into tissue culture cells, then the cells were treated with nocodazole to depolymerize the spindle. Cells that received the hsMAD2 antibodies failed to arrest in the presence of nocodazole. The kinetochore-localization domain of mBub1 resides in its amino terminus. Taylor and McKeon [7] overexpressed the this amino-terminal mBub1 domain in tissue culture cells. The overexpressing cells arrested poorly compared to wild-type cells in the presence of nocodazole. Overexpression of full-length Bub1 in yeast confers a similar phenotype, causing cells to become hypersensitive to spindle depolymerization [21]. It is likely that Bub1 overexpression disrupts the ability of the kinetochore to signal checkpoint arrest in the absence of a spindle. Checkpointarrested tissue culture cells eventually recover from the arrest and die, exhibiting characteristics of apoptosis (Figure 1, adaptation). In contrast, cells that express the amino-terminal mBub1 fragment lack the checkpoint, continue into the next cell cycle, and become polyploid without signs of apoptotic elimination. The spindle assembly checkpoint may serve a dual role in mediating arrest so that the cell can correct errors, and in the elimination of damaged cells. R615 Does the lack of a spindle assembly checkpoint affect the timing of a normal cell cycle? Taylor and McKeon [7] investigated this question by examining the effect of overexpressing the mBub1 amino-terminal domain on the timing of the cell cycle. They found that the overexpression had no effect on the timing of G1, S or G2 phases of the cell cycle, but delayed the exit from mitosis by approximately 25 minutes. Yeast cells that lack the spindle assembly checkpoint protein Mad1 are also accelerated in their exit from mitosis (P. Dann and A. Rudner, personal communication). Thus, the spindle checkpoint may have a role during the normal cell cycle in restraining the passage through mitosis. Other proteins involved in the spindle assembly checkpoint have been found to be resident components of the kinetochore. Studies with budding yeast have shown that the Mad2 protein interacts tightly with another spindle assembly checkpoint protein, Mad1, and studies with Xenopus extracts have shown that XMAD2 interacts with a Mad1-like protein (R. Chen and K. Hardwick, personal communication). The Mad1 protein has also been shown to be phosphorylated by the protein kinase Mps1, and overexpression of Mps1 causes constitutive activation of the spindle assembly checkpoint [22]. The Bub1 protein kinase also interacts with another checkpoint protein, Bub3, forming an active kinase complex [21]. Thus, the kinetochore may contain as many as five proteins directly involved in the spindle assembly checkpoint, two of which are protein kinases. Integrating information at the kinetochore with the cell cycle engine Exit from mitosis is normally mediated by ubiquitin-dependent proteolysis of B-type cyclins, resulting in a drop in cyclin-dependent kinase levels [23]. Activation of the spindle assembly checkpoint blocks exit from mitosis, arresting cells with high B-type cyclin levels and elevated cyclin-dependent kinase activity [20,24]. The question remains as to how the kinetochore generates the signal to arrest the cell cycle, and how that signal is transduced to the cell cycle machinery. Studies with X. laevis extracts have shown that a well known component of kinase cascades, the mitogen-activated protein (MAP) kinase ERK-2, is required for both the establishment and maintenance of the spindle assembly checkpoint [20]. In addition, studies with yeast have shown that mutations in a B-type subunit of a type-II protein phosphatase cause a spindle assembly checkpoint phenotype [24,25]. Recent experiments suggest that the inhibitory signal for checkpoint arrest may not be mediated by a soluble factor, because the signal is not transferred between two spindles in the same cytoplasm [26]. One of the most attractive targets of the checkpoint signal is the cyclososme/anaphase-promoting complex (APC), the enzyme that ubiquitinates cyclins, targeting them for R616 Current Biology, Vol 7 No 10 destruction [23]. Cyclin ubiquitination activity is inhibited in extracts from checkpoint-arrested yeast cells, but it is not known whether this is a direct or indirect effect of the checkpoint on the cyclosome/APC [27]. Other targets, such as the proteasome itself or the hydrolases that remove ubiquitin from proteins, are still potential candidates for regulation. Although the signalling pathway from the kinetochore to the cell cycle engine is unknown, current studies suggest that checkpoint proteins on kinetochores are able to sense the attachment of the chromosomes to the spindle microtubules and, in response to improper attachment, delay the cell cycle in order to promote faithful chromosome segregation. References 1. Rudner AD, Murray AW: The spindle assembly checkpoint. Curr Opin Cell Biol 1996, 8:773-780. 2. Wells WAE: The spindle-assembly checkpoint: aiming for a perfect mitosis, every time. Trends Cell Biol 1996, 6:228-234. 3. Li R, Murray AW: Feedback control of mitosis in budding yeast. Cell 1991, 66:519-531. 4. Li Y, Benezra R: Identification of a human mitotic checkpoint gene: hsMAD2. Science 1996, 274:246-248. 5. Hoyt MA, Trotis L, Roberts BT: S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 1991, 66:507-517. 6. Chen R-H, Waters JC, Salmon ED, Murray AW: Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science 1996, 274:242-246. 7. Taylor SS, McKeon F: Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 1997, 89:727-735. 8. Wells WAE, Murray AW: Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol 1996, 133:75-84. 9. Brues AM, Cohen A: Effects of colchicine and related substances on cell division. Biochem J 1936, 30:1363. 10. Brues AM, Jackson EB: Nuclear abnormalities resulting from inhibition of mitosis by colchicine and other substances. Am J Cancer 1937, 30:504-511. 11. Rieder CL, Schultz A, Cole R, Sluder G: Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol 1994, 127:1301-1310. 12. Rieder CL, Cole RW, Khodjakov A, Sluder G: The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 1995, 130:941-948. 13. Clayton L, Pogson CI, Gull K: Microtubule proteins in the yeast, Saccharomyces cerevisiae. FEBS Lett 1979, 106:67-70. 14. Merdes A, De Mey J: The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur J Cell Biol 1990, 53:313-325. 15. Hayden JH, Bowser SS, Rieder CL: Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J Cell Biol 1990, 111:1039-1045. 16. Ault JG, Nicklas RB: Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma 1989, 98:33-39. 17. Nicklas RB, Ward SC: Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol 1994, 126:1241-1253. 18. Rieder CL, Salmon ED: Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol 1994, 124:223-233. 19. Nicklas RB: How cells get the right chromosomes. Science 1997, 275:632-637. 20. Minshull J, Sun H, Tonks NK, Murray AW: MAP-kinase dependent mitotic feedback arrest in Xenopus egg extracts. Cell 1994, 79:475-486. 21. Roberts RT, Farr KA, Hoyt MA: The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol 1994, 14:8282-8291. 22. Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW: Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 1996, 273:953-956. 23. King RW, Deshaies RJ, Peters JM, Kirschner MW: How proteolysis drives the cell cycle. Science 1996, 274:1652-1659. 24. Minshull J, Straight A, Rudner A, Dernburg A, Belmont A, Murray AW: Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol 1996, 6:1609-1620. 25. Wang Y, Burke DJ: Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol 1997, 17:620-626. 26. Rieder CL, Khodjakov A, Paliulis LV, Fortier TM, Cole RW, Sluder G: Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc Natl Acad Sci USA 1997, 94:5107-5112. 27. Zachariae W, Nasmyth K: TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell 1996, 7:791-801.