* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download File - Integrated Science
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genetic engineering wikipedia , lookup
Metagenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
Human genome wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transposable element wikipedia , lookup
Hammerhead ribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Microevolution wikipedia , lookup
Genetic code wikipedia , lookup
History of genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Messenger RNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Polyadenylation wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Primary transcript wikipedia , lookup
Epitranscriptome wikipedia , lookup
History of RNA biology wikipedia , lookup
RNA-binding protein wikipedia , lookup
Non-coding RNA wikipedia , lookup
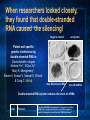
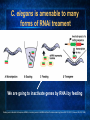
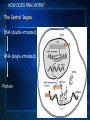
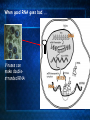
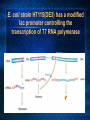
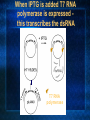
Introduction to C. elegans and RNA interference Today’s Goals Review RNAi How it works to silence genes in C. elegans How we will use it in the lab Prepare RNAi The problem In order to understand biology, we need to learn about the function of the underlying genes How can we find out what genes do? One way is by eliminating the functional protein, and examining the phenotype Called reverse genetics Some approaches to Reverse Genetics Targeted deletion by homologous recombination (KNOCKOUT MICE) - We’ll discuss next time Specific mutational changes can be made Time consuming and limited to certain organisms Antisense RNA Variable effects and mechanism not understood With the completion of the genome sequencing project, a quicker, less expensive reverse genetics method was needed. Luckily scientists discovered . . . RNAi The old model: Antisense RNA leads to translational inhibition mRNA is considered the sense strand antisense RNA is complementary to the sense strand The old model: Antisense RNA leads to translational inhibition This can give the same phenotype as a mutant An experiment showed that the antisense model didn’t make sense: The antisense technology was used in worms Puzzling results were produced: both sense and antisense RNA preparations were sufficient to cause interference. What could be going on? 1995 Guo S, and Kemphues KJ. First noticed that sense RNA was as effective as antisense RNA for suppressing gene expression in worm When researchers looked closely, they found that double-stranded RNA caused the silencing! Negative control uninjected Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans Andrew Fire*, SiQun Xu*, Mary K. Montgomery*, Steven A. Kostas*†, Samuel E. Driver‡ & Craig C. Mello‡ mex-3B antisense RNA mex-3B dsRNA Double-stranded RNA injection reduces the levels of mRNA 1998 Fire et al. First described RNAi phenomenon in C. elegans by injecting dsRNA into C. elegans which led to an efficient sequencespecific silencing and coined the term "RNA Interference". C. elegans is amenable to many forms of RNAi treament We are going to inactivate genes by RNAi by feeding Feeding worms bacteria that express dsRNAs or soaking worms in dsRNA sufficient to induce silencing (Gene 263:103, 2001; Science 282:430, 1998) HOW DOES RNAi WORK? The Central Dogma DNA (double-stranded) RNA (single-stranded) Protein When good RNA goes bad.... Viruses can make doublestranded RNA When good RNA goes bad.... Cells sense doublestranded RNA and activate a response called RNAi Understanding how RNAi works is the key to using it as a genetic tool and for therapy Dicer cuts dsRNA into short RNAs Vanhecke, D.; Janitz, M. Drug Discov. Today, 2005, 10, 205-225. Role for a bidentate ribonuclease in the initiation step of RNA interference Emily Bernstein, Amy A. Caudy, Scott M. Hammond & Gregory J. Hannon 2001 Bernstein et al. Cloned Dicer, the RNase III enzyme that is evolutionarily conserved and contains helicase and PAZ domains, as well as two dsRNA-binding domains. How does the RNAi that we are using deliver dsRNA to the worms? We are going to feed our worms a bacterial strain that has been engineered to express a dsRNA using a plasmid vector Silencinggenomes.org supplies the bacterial strain with the recombinant plasmid, already ready to use! How does the vector induce RNAi? • Allows E. coli to produce double-stranded RNA • Requires a specific strain of E. coli The RNAi feeding vector has two T7 RNA polymerase promoters • T7 RNA polymerase is a viral polymerase. • It binds to a specific T7 promoter sequence. • The L4440 vector is designed with two T7 promoters to make dsRNA. E. coli strain HT115(DE3) has a modified lac promoter controlling the transcription of T7 RNA polymerase T7 RNA polymerase T7 RNA polymerase When IPTG is added T7 RNA polymerase is expressed this transcribes the dsRNA T7 RNA polymerase Summary RNAi is a fast and efficient way to silence gene expression and investigate gene function The RNAi we will perform uses genetically engineered bacteria that express dsRNA by induction using IPTG in the agar, and feeding of the bacteria to the C. elegans