* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bacterial Regulation
Non-coding DNA wikipedia , lookup
Magnesium transporter wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Messenger RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Molecular evolution wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding RNA wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Epitranscriptome wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Point mutation wikipedia , lookup
Expression vector wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
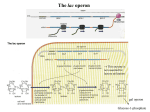
PowerPoint Presentation Materials to accompany Genetics: Analysis and Principles Robert J. Brooker CHAPTER 14 GENE REGULATION IN BACTERIA AND BACTERIOPHAGES Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display INTRODUCTION The term gene regulation means that the level of gene expression can vary under different conditions Genes that are unregulated are termed constitutive They have essentially constant levels of expression Frequently, constitutive genes encode proteins that are necessary for the survival of the organism The benefit of regulating genes is that encoded proteins will be produced only when required Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-2 INTRODUCTION Gene regulation is important for cellular processes such as 1. Metabolism 2. Response to environmental stress 3. Cell division Regulation can occur at any of the points on the pathway to gene expression Refer to Figure 14.1 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-3 Figure 14.1 14-4 14.1 TRANSCRIPTIONAL REGULATION The most common way to regulate gene expression in bacteria is at the transcriptional level The rate of RNA synthesis can be increased or decreased Negative control: Repressors Bind to DNA and inhibit transcription Positive control: Activators Bind to DNA and increase transcription Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-5 In some cases, the presence of a small effector molecule may increase transcription These molecules are termed inducers Bind activators and cause them to bind to DNA Bind repressors and prevent them from binding to DNA In other cases, the presence of a small effector molecule may inhibit transcription Corepressors bind to repressors and cause them to bind to DNA Inhibitors bind to activators and prevent them from binding to DNA Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-6 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Promoter Fig. 14.2ab(TE Art) DNA Repressor protein No transcription RNA polymerase or Transcription occurs Inducer Repressor protein Repressor protein, inducer molecule, inducible gene RNA polymerase Transcription occurs Repressor protein or Corepressor Repressor protein No transcription Repressor protein, corepressor molecule, repressible gene Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. transcription or Art) Fig.No14.2cd(TE Activator protein Inducer RNA polymerase Activator protein Transcription occurs Activator protein, inducer molecule, inducible gene RNA polymerase Activator protein Transcription or occurs No transcription Inhibitor Activator protein Activator protein, inhibitor molecule, repressible gene The Phenomenon of Enzyme Adaptation At the turn of the 20th century, scientists made the following observation A particular enzyme appears in the cell only after the cell has been exposed to the enzyme’s substrate This observation became known as enzyme adaptation François Jacob and Jacques Monod at the Pasteur Institute in Paris were interested in this phenomenon They focused their attention on lactose metabolism in E. coli to investigate this problem Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-9 The lac Operon An operon is a regulatory unit consisting of a few structural genes under the control of one promoter It encodes polycistronic mRNA that contains the coding sequence for two or more structural genes This allows a bacterium to coordinately regulate a group of genes that encode proteins with a common function Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-10 Figure 16-3 Copyright © 2006 Pearson Prentice Hall, Inc. An operon contains several different regions There are two distinct transcriptional units 1. The actual lac operon a. DNA elements Promoter Binds RNA polymerase Operator Binds the lac repressor protein CAP site Binds the Catabolite Activator Protein (CAP) b. Structural genes lacZ Encodes b-galactosidase Enzymatically cleaves lactose and lactose analogues Also converts lactose into allolactose (an isomer) lacY Encodes lactose permease Membrane protein required for transport of lactose and analogues lacA Encodes transacetylase Covalently modifies lactose and analogues Its functional necessity remains unclear Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-11 Figure 14.3 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-13 Figure 14.3a shows the organization and transcriptional regulation of the lac operon genes There are two distinct transcriptional units 2. The lacI gene Not considered part of the lac operon Has its own promoter, the i promoter Constitutively expressed at fairly low levels Encodes the lac repressor The lac repressor protein functions as a tetramer Only a small amount of protein is needed to repress the lac operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-12 The lac Operon Is Regulated By a Repressor Protein The lac operon can be transcriptionally regulated 1. By a repressor protein 2. By an activator protein The first method is an inducible, negative control mechanism It involves the lac repressor protein The inducer is allolactose It binds to the lac repressor and inactivates it Refer to Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-14 RNA pol cannot access the promoter Constitutive expression The lac operon is now repressed Therefore no allolactose Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-15 Translation The lac operon is now induced The conformation of the repressor is now altered Repressor can no longer bind to operator Some gets converted to allolactose Figure 14.4 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-16 Figure 16-5 Copyright © 2006 Pearson Prentice Hall, Inc. Repressor does not completely inhibit transcription So very small amounts of the enzymes are made Figure 14.5 The cycle of lac operon induction and repression Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-17 Experiment 14A: The lacI Gene Encodes a Repressor Protein In the 1950s, Jacob and Monod, and their colleague Arthur Pardee, had identified a few rare mutant strains of bacteria with abnormal lactose adaptation One type of mutant involved a defect in the lacI gene It was designated lacI– It resulted in the constitutive expression of the lac operon even in the absence of lactose The lacI– mutations mapped very close to the lac operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-18 Jacob, Monod and Pardee proposed two different functions for the lacI gene Figure 14.6 Jacob, Monod and Pardee applied a genetic approach to distinguish between the two hypotheses Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-19 They used bacterial conjugation methods to introduce different portions of the lac operon into different strains They identified F’ factors (plasmids) that carried portions of the lac operon For example: Consider an F’ factor that carries the lacI gene Bacteria that receive this will have two copies of the lacI gene One on the chromosome and the other on the F’ factor These are called merozygotes, or partial diploids Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-20 Merozygotes were instrumental in allowing Jacob et al to elucidate the function of the lacI gene There are two key points 1. The two lacI genes in a merozygote may be different alleles lacI– on the chromosome lacI+ on the F’ factor 2. Genes on the F’ factor are not physically connected to those on the bacterial chromosome If hypothesis 1 is correct The repressor protein produced from the F’ factor can diffuse and regulate the lac operon on the bacterial chromosome If hypothesis 2 is correct The binding site on the F’ factor cannot affect the lac operon on the bacterial chromosome, because they are not physically adjacent Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-21 The Hypothesis The lacI gene either/or 1. Encodes a regulatory protein (the lac repressor) that can diffuse throughout the cell 2. Acts as a binding site for a repressor protein Thus it can only act on a physically connected lac operon Testing the Hypothesis Refer to Figure 14.7 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-22 Figure 14.7 14-23 Figure 14.7 14-24 Figure 14.7 14-25 The Data Strain Addition of lactose Amount of b-galactosidase (percentage of parent strain) Mutant No 100% Mutant Yes 100% Merozygote No <1% Merozygote Yes 220% Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-26 Interpreting the Data Strain Addition of lactose Expected result because of constitutive expression in the lacI– strain Amount of b-galactosidase (percentage of parent strain) Mutant No 100% Mutant Yes 100% Merozygote No <1% Merozygote Yes 220% In the presence of lactose, both lac operons are induced, yielding a higher level of enzyme activity In the absence of lactose, both lac operons are repressed This result is consistent with hypothesis 1 The lacI gene codes for a repressor protein that can diffuse throughout the cell and bind to any lac operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-27 Interpreting the Data The interaction between regulatory proteins and DNA sequences have led to two definitions 1. Trans-effect Genetic regulation that can occur even though DNA segments are not physically adjacent Mediated by genes that encode regulatory proteins Example: The action of the lac repressor on the lac operon 2. Cis-effect or cis-acting element A DNA sequence that must be adjacent to the gene(s) it regulates Mediated by sequences that bind regulatory proteins Example: The lac operator Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-28 Interpreting the Data Table 14.1 summarizes the effects of lacI gene mutations versus lacO (operator) in merozygotes Overall A mutation in a trans-acting factor is complemented by the introduction of a second gene with a normal function However, a mutation in a cis-acting element is not! Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-29 Figure 16-6 Copyright © 2006 Pearson Prentice Hall, Inc. Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-30 Figure 16-7 Copyright © 2006 Pearson Prentice Hall, Inc. Table 16-1 Copyright © 2006 Pearson Prentice Hall, Inc. The lac Operon Is Also Regulated By an Activator Protein The lac operon can be transcriptionally regulated in a second way, known as catabolite repression When exposed to both lactose and glucose E. coli uses glucose first, and catabolite repression prevents the use of lactose When glucose is depleted, catabolite repression is alleviated, and the lac operon is expressed Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-31 The lac Operon Is Also Regulated By an Activator Protein The small effector molecule in catabolite repression is not glucose This form of genetic regulation involves a small molecule, cyclic AMP (cAMP) It is produced from ATP via the enzyme adenylyl cyclase cAMP binds an activator protein known as the Catabolite Activator Protein (CAP) Also termed the cyclic AMP receptor protein (CRP) Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-32 Figure 16-9 Copyright © 2006 Pearson Prentice Hall, Inc. The lac Operon Is Also Regulated By an Activator Protein The cAMP-CAP complex is an example of genetic regulation that is inducible and under positive control The cAMP-CAP complex binds to the CAP site near the lac promoter and increases transcription In the presence of glucose, the enzyme adenylyl cyclase is inhibited This decreases the levels of cAMP in the cell Therefore, cAMP is no longer available to bind CAP Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-33 Figure 18.23 Positive control of the lac operon by catabolite activator protein (CAP) Promoter DNA lacl lacZ CAP-binding site Active CAP cAMP Inactive CAP RNA polymerase can bind and transcribe Operator Inactive lac repressor (a) Lactose present, glucose scarce (cAMP level high): abundant lac mRNA synthesized. If glucose is scarce, the high level of cAMP activates CAP, and the lac operon produces large amounts of mRNA for the lactose pathway. Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Promoter DNA lacl lacZ CAP-binding site Operator RNA polymerase can’t bind Inactive CAP Inactive lac repressor (b) Lactose present, glucose present (cAMP level low): little lac mRNA synthesized. When glucose is present, cAMP is scarce, and CAP is unable to stimulate transcription. Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Both negative and positive control mechanisms are used by the cell. The lac Operon Has Three Operator Sites for the lac Repressor Detailed genetic and crystallographic studies have shown that the binding of the lac repressor is more complex than originally thought In all, three operator sites have been discovered O1 Next to the promoter O2 Downstream in the lacZ coding region O3 Slightly upstream of the CAP site Refer to Figure 14.9 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-36 Repression is 1,300 fold Therefore, transcription is 1/1,300 the level when lactose is present No repression ie: Constitutive expression Figure 14.9 The identification of three lac operator sites 14-37 The results of Figure 14.9 supported the hypothesis that the lac repressor must bind to two of the three operators to cause repression It can bind to O1 and O2 , or to O1 and O3 But not O2 and O3 If either O2 or O3 is missing maximal repression is not achieved Binding of the lac repressor to two operator sites requires that the DNA form a loop A loop in the DNA brings the operator sites closer together This facilitates the binding of the repressor protein Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-38 Each repressor dimer binds to one operator site Each repressor dimer binds to one operator site Figure 14.10 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-39 The ara Operon Another operon in E. coli that is involved in sugar metabolism is the ara (arabinose) operon It contains Three structural genes involved in arabinose metabolism These are designated araB, araA and araD A single promoter, PBAD A CAP site, which binds the catabolite activator protein Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-40 The araC gene is adjacent to the ara operon It has its own promoter, PC It encodes a regulatory protein, AraC AraC can bind to three different operator sites Designated araI, araO1 and araO2 The AraC protein can act as either a negative or positive regulator of transcription Depending on whether or not arabinose is present Figure 14.11 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-41 AraC protein binds to all three operators AraC dimer bound to araO1 inhibits transcription of the araC gene This keeps AraC protein levels fairly low AraC monomers bound to araO2 and araI repress the ara operon They bind to each other (via looped DNA), and block RNA pol access to PBAD araO1 Figure 14.12 14-42 Arabinose binds to the AraC proteins The interaction betweem the AraC proteins at the araO2 and araI is broken This breaks the DNA loop Another, AraC protein binds to araI This AraC dimer at araI activates transcription CAP-cAMP activation occurs when glucose levels are low Figure 14.12 14-43 Figure 16-15 Copyright © 2006 Pearson Prentice Hall, Inc. Regulation of a anabolic pathway (a) Regulation of enzyme activity Precursor Feedback inhibition Enzyme 1 Enzyme 2 Enzyme 3 (b) Regulation of enzyme production Gene 1 Gene 2 Regulation of gene expression Gene 3 – Enzyme 4 Gene 4 – Enzyme 5 Tryptophan Copyright © 2005 Pearson Education, Inc. publishing as Benjamin Cummings Gene 5 The trp Operon The trp operon (pronounced “trip”) is involved in the biosynthesis of the amino acid tryptophan The genes trpE, trpD, trpC, trpB and trpA encode enzymes involved in tryptophan biosynthesis The genes trpR and trpL are involved in regulation trpR Encodes the trp repressor protein Functions in repression trpL Encodes a short peptide called the Leader peptide Functions in attenuation Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-44 RNA pol can bind to the promoter Cannot bind to the operator site Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-45 Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-46 Another mechanism of regulation Figure 14.13 Organization of the trp operon and regulation via the trp repressor protein 14-47 Attenuation occurs in bacteria because of the coupling of transcription and translation During attenuation, transcription actually begins but it is terminated before the entire mRNA is made A segment of DNA, termed the attenuator, is important in facilitating this termination In the case of the trp operon, transcription terminates shortly past the trpL region (Figure 14.13c) Thus attenuation inhibits the further production of tryptophan The segment of trp operon immediately downstream from the operator site plays a critical role in attenuation The first gene in the trp operon is trpL It encodes a short peptide termed the Leader peptide Refer to Figure 14.14 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-48 Region 2 is complementary to regions 1 and 3 Region 3 is complementary to regions 2 and 4 Therefore several stem-loops structures are possible The 3-4 stem loop is followed by a sequence of Uracils It acts as an intrinsic terminator These two codons provide a way to sense if there is sufficient tryptophan for translation Figure 14.14 Sequence of the trpL mRNA produced during attenuation 14-49 Formation of the 3-4 stem-loop causes RNA pol to terminate transcription at the end of the trpL gene Conditions that favor the formation of the 3-4 stem-loop rely on the translation of the trpL mRNA There are three possible scenarios 1. No translation 2. Low levels of tryptophan 3. High levels of tryptophan Refer to Figure 14.15 Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-50 Transcription terminates just past the trpL gene Most stable form of mRNA occurs Therefore no coupling of transcription and translation Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-51 Region 1 is blocked 3-4 stem-loop does not form RNA pol transcribes rest of operon Insufficient amounts of tRNAtrp Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-52 Sufficient amounts of tRNAtrp 3-4 stem-loop forms Translation of the trpL mRNA progresses until stop codon RNA polymerase pauses Transcription terminates Region 2 cannot base pair with any other region Figure 14.15 Possible stem-loop structures formed from trpL mRNA under different conditions of translation 14-53 Inducible vs Repressible Regulation The study of many operons revealed a general trend concerning inducible versus repressible regulation Operons involved in catabolism (ie. breakdown of a substance) are typically inducible The substance to be broken down (or a related compound) acts as the inducer Operons involved in anabolism (ie. biosynthesis of a substance) are typically repressible The inhibitor or corepressor is the small molecule that is the product of the operon Copyright ©The McGraw-Hill Companies, Inc. Permission required for reproduction or display 14-54