* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 20141203103493
RNA interference wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transcription factor wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
RNA silencing wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Protein moonlighting wikipedia , lookup
Polyadenylation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular evolution wikipedia , lookup
Messenger RNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Proteolysis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Point mutation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene regulatory network wikipedia , lookup
Two-hybrid screening wikipedia , lookup
List of types of proteins wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression wikipedia , lookup
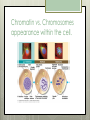
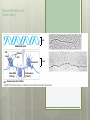
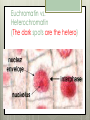
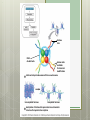
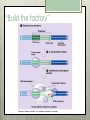
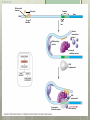
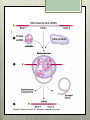
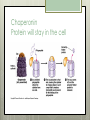
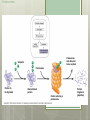
AP Biology Eukaryotic Genome Control Mechanisms for Gene expression Chromatin vs. Chromosomes appearance within the cell. Histone Proteins and Supercoiling 2 nm DNA double helix Histones Histone tails Histone H1 Linker DNA (“string”) Nucleosomes (10-nm fiber) Nucleosome (“bead”) 10 nm Supercoiling of Chromatin Histones-proteins DNA wraps around Nucleosome-unit of DNA wrapped around histones Supercoiling-Chromatinchromosomes Heterochromatin-remains condensed Euchromatin-loose during interphase Cellular differentiation-making cells different; accomplished by turning genes “on” or “off”differential gene expression Euchromatin vs. Heterochromatin (The dark spots are the hetero) During Transcription DNA Methylation-heavy coat-preventssource of genomic imprinting Histone acetylation-breaks bond btwn DNA and proteins;allows RNA polymerase to attach piwi-associated RNAs(piRNAs) induce formation of heterochromatin Building transcription initiation complex Enhancers& activators-control rate Repressor or silencer sit on TATA box Signal NUCLEUS Chromatin DNA Gene available for transcription Gene Transcription RNA DNA Control stages in Protein Synthesis Exon Primary transcript Intro RNA processing Tail Cap mRNA in nucleus Transport to cytoplasm CYTOPLASM mRNA in cytoplasm Degradation of mRNA Translation Polypeptide Cleavage Chemical modification Transport to cellular destination Active protein Degradation of protein Degraded protein Methylation . Histone tails DNA double helix Amino acids available for chemical modification Histone tails protrude outward from a nucleosome Unacetylated histones Acetylated histones Acetylation of histone tails promotes loose chromatin structure that permits transcription “Build the factory” Enhancers Distal control element Activators Promoter Gene DNA TATA box Enhancer General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II RNA polymerase II Transcription Initiation complex RNA synthesis Liver cell nucleus Notice the different nucleotide control sequences (red vs, pink) Available activators Enhancer Control elements Lens cell nucleus Available activators Promoter Albumin gene Crystallin gene Albumin gene not expressed Albumin gene expressed Crystallin gene not expressed Liver cell Crystallin gene expressed Lens cell miRNA &siRNA Protein complex Degradation of mRNA Dicer OR miRNA Target mRNA Hydrogen bond Blockage of translation Coordinated control of genes familiesmultiple copies of same gene, all transcribed at one MicroRNA (miRNA) and small interfering RNA (siRNA)-little pieces that attach to mRNA to control transcription Post transcription Regulation Alternative RNA Splicing using snRPS Cytoplasmic degradation Control of exons Translation control mechanisms Build Translation Initiation Complex Faulty cap Tail too short How many As on the tail? “Build the factory” Post Translation control Chaperonin or SRP Phosphorylation-turns on or off Proteosomes-control how long protein lasts-mainly work on intracellularly produced proteins ex. Cyclins, remove transcription factors, recycle amino acids (Lysosomes work on extracellularly produced proteins) Chaperonin Protein will stay in the cell RER Protein will leave the cell Proteosomes Proteasome and ubiquitin to be recycled Ubiquitin Proteasome Protein to be degraded Ubiquitinated protein Protein entering a proteasome Protein fragments (peptides) Nuclear membrane’s role? What is Dark Matter? A five-year project called ENCODE, for "Encyclopedia of DNA Elements," found that about 80 percent of the human genome is biologically active, influencing how nearby genes are expressed and in which types of cells. It's not junk DNA, which was previously thought — instead, these non-coding regions of DNA could have major bearing on diseases and genetic mutations, researchers say. Epigenetic Inheritance of traits not directly involving nucleotide sequence Modifications of chromatin can be reversed Methylation ID twins: one normal; one schizophrenic