* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Epigenetics - Cayetano Heredia University
Saethre–Chotzen syndrome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Human genome wikipedia , lookup
Genome evolution wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Non-coding DNA wikipedia , lookup
Primary transcript wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Epigenetic clock wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Designer baby wikipedia , lookup
Y chromosome wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
DNA methylation wikipedia , lookup
Oncogenomics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Microevolution wikipedia , lookup
Epigenomics wikipedia , lookup
Genome (book) wikipedia , lookup
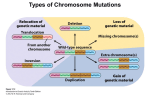
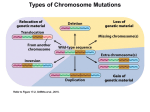
Point mutation wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Neocentromere wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
OBG420 Janine LaSalle, Ph.D. Epigenetics, Parental Imprinting, and X Inactivation Epigenetics Inherited and reversible modifications to nucleotides or chromosomes that do not change the sequence but can alter gene expression Epigenetics • Epigenetics can explain how multicellular organisms can express different genes in different tissues using the same set of chromosomes. • Epigenetics can explain early environmental influences on eventual phenotype. • Epigenetics can also explain non-Mendelian modes of inheritance such as genetic imprinting. PARENTAL IMPRINTING THE CONTRIBUTION OF BOTH PARENTS IS REQUIRED FOR NORMAL EMBRYONIC DEVELOPMENT IN MAMMALS EGG SPERM EGG EGG SPERM SPERM TRANSPLANT FERTILIZED EGG WITHOUT NUCLEUS EMBRYO MOUSE GYNOGENETIC ANDROGENETIC FAILURE OF EXTRAEMBRYONIC DEVELOPMENT FAILURE OF EMBRYONIC DEVELOPMENT Human Parthenogenetic Tumors • Hydatidiform moles – Androgenetic placental tumor • Ovarian teratoma – Gynogenetic benign tumor Parental Imprinting of 15q11-13 Normal M P Chromosome 15 Prader-Willi Syndrome M P M M M del Paternal Deletion Angelman Syndrome P P P M P del Maternal Disomy Maternal Deletion Paternal Disomy Maternal Inheritence Inheritance Uniparental Disomy (UPD) of Chromosome 15 in Prader-Willi (PWS) and Angelman Syndromes (AS) Chromosome loss PWS Maternal heterodisomy Chromosome duplication Meiosis I nondisjunction Reciprocal nondisjunction products Fertilization with normal sperm AS Paternal isodisomy Chromosome UPD in humans MATERNAL PATERNAL NO PHENOTYPE 15 PWS 15 AS 13 6 IUGR 11 BWS 17 7 IUGR 14 MR 21 16 IUGR 21 22 IUGR 14 GR IUGR Intrauterine growth retardation Growth retardation 20 GR GR BWS AS PWS Beckwith-Weidemann syndrome Angelman syndrome Prader-Willi syndrome Characteristic features of imprinted loci • Allele specific differences in: – Transcription – Methylation – DNA replication Parental Imprinting Allele-specific Transcription • Transcription is silenced or imprinted on one parental allele – ex) H19 is paternally imprinted, expressed only from maternal allele • Imprinted monoalleleic expression can be in all tissues (SNRPN, H19) or limited to certain tissues (UBE3A) or developmental stages (INS) Parental Imprinting Allele-specific Transcription Paternal specific expression of SNRPN DNA DNA RNA DNA Examples of Imprinted Genes Chromosomal region 15q11-13 Maternal Imprint SNRPN, IPW, NDN, PAR1, PAR5 Paternal Imprint UBE3A 11p15.5 IGF2 H19, p57KIP KvLQT1, IPL 11p13 6q26-27 7q32 19q13.4 20q13 Xq13.2 WT1 IGF2R/M6P PEG1/MEST PEG3 GNAS1 XIST Parental Imprinting Allele-specific Methylation • A covalent modification to cytosine in higher eukaryotes (animals, plants, fungi) Parental Imprinting Allele-specific Methylation • CpG (5'-C-G-3') occurs less frequently in the mammalian genome than any other dinucleotide. • CpG islands are clusters of CpG sites near the 5' ends of genes and are the target of DNA methylation. What role does methylation have on gene expression? • Usually results in gene silencing, but not always • Multiple methylation sites at the 5’ end and within introns of a gene may regulate transcription • Methylation may involved in determining and/or maintaining genes in inactive state Parental Imprinting Allele-specific Methylation • Parental allele-specific methylation found around imprinted genes • Erasure and re-establishment in gametes for each generation • Some methylation imprints are inherited from gametes, but most are erased and reestablished in early development Parental Imprinting Allele-specific Methylation Parental Imprinting Allele-specific DNA Replication • Most homologous chromosomal regions replicate synchronously in S phase • Parental allele-specific asynchronous replication of imprinted regions • Asynchronous replication is established in the gametes and maintained throughout development Parental Imprinting Allele-specific DNA Replication Theories for parental imprinting • Parental “tug of war” in evolution – Igf2 is paternally expressed, Igf2r is maternally expressed • Protection against trophoblastic disease – Lack of trophoblast in parthenogenetic embryos – Molar pregnancy, high rate of malignancy • Biparental exchange or cross-talk – Different replication timing of UPDs – Homologous association Parental Imprinting and Mammalian Reproductive Cloning • Many cloned livestock exhibit “large offspring syndrome” due to dysregulated expression of Igf2. • Cloned mice and ES cells have many epigenetic defects in imprinted genes. • Human children from in vitro fertilization (IVF) have increased rates of Angelman and Beckwith-Wiedemann syndromes. X chromosome inactivation In females (46,XX), X inactivation serves to reduce the amount of transcriptionally active X chromatin in somatic cells. In female somatic cells, one X chromosome becomes inactive and is cytologically detected as a Barr body. X inactivation • The number of X chromosomes are counted prior to X inactivation. • X inactivation follows the "n-1" rule so that only one X chromosome remains active in each cell, regardless of X chromosome copy number. X inactivation X inactivation is mediated by transcription of the gene XIST from the X inactivation center (XIC). XIST is only expressed from the inactive X chromosome and does not encode a protein but rather functions as an RNA molecule. X inactivation: Role of XIST X inactivation • The inactive X chromosome in female cells is more heavily methylated and later replicating than the active X chromosome • There are pseudoautosomal regions of the X chromosome that are transcriptionally active on both active and inactive X chromosomes. Random parental X inactivation in somatic cells Around the stage of implantation, there is an erasure and re-establishment of X inactivation patterns. X inactivation in females becomes random, meaning that roughly 50% of the cells have inactivated the paternal X chromosome and the other half are inactive on the maternal X chromosome. Random parental X inactivation in somatic cells Random parental X inactivation in somatic cells • Female somatic cells can be mosaic for expression of alleles or mutations of a gene. Non-random X inactivation can be observed in some women and can be associated with increased frequency of spontaneous abortion. • Female carriers of X-linked disorders often show skewed or non-random X inactivation patterns. Human diseases involving methylation Rett Syndrome Rett syndrome is an X-linked dominant disorder affecting heterozygous females. Rett syndrome infants develop normally until 6 to 18 months of age but then develop a progressive loss of neurodevelopmental milestones. Mutations in the methylation-specific binding protein MECP2 on the X chromosome cause Rett syndrome. Human diseases involving methylation Rett Syndrome Females are heterozygous and mosaic for expression of MECP2 mutations in somatic cells. MECP2 mutation in males is rare because mutations are more frequent on paternal allele. MECP2 mutation in males can cause MR. Some cases of inheritance from a female carrier have been reported, in which the phenotypically normal female carrier shows non-random Xinactivation. Human diseases involving methylation Rett Syndrome Methylated DNA is recognized by methylationspecific binding proteins such as MeCP2. • MeCP2 localizes DNA to transcriptionally inactive heterochromatin and associates with proteins involved in histone packaging and transcriptional silencing. Human diseases involving methylation ICF Syndrome Variable immunodeficiency centromeric heterochromatin instability and facial abnormalities • Cytogenetics reveals many chromosomal breaks and interchanges • Hypomethylation of centromeric satellite repeats results in instability of chromosomes during mitosis • Mutations were found in ICF patients within the DNMT3B gene encoding a DNA methyltransferase Human diseases involving methylation Fragile X Syndrome Expansion of CGG triplet repeat in the 5’ untranslated region of FMR1 on X chromosome • Pre-mutation (50-230 repeats) can be expanded to full mutation (>230 repeats) by passage through a female carrier • Hypermethylation of the expanded CGG repeat can result is loss of expression of the FMR-1 gene Why does methylation exist? • Genome defense mechanism – Foreign DNA becomes methylated in order to silence undesired transcription and reduce recombination • Developmental control of gene expression – Imprinting in mammals, meiosis in fungi • Development of the central nervous system in mammals – Multiple human neurodevelopmental disorders have epigenetic etiologies