* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download the ubiquitin system and a putative stimulatory role
Molecular evolution wikipedia , lookup
Genetic code wikipedia , lookup
Endomembrane system wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Magnesium transporter wikipedia , lookup
Gene regulatory network wikipedia , lookup
Metalloprotein wikipedia , lookup
Acetylation wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Expression vector wikipedia , lookup
Interactome wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
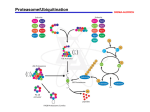
The ubiquitin system and a putative stimulatory role for activator turnover in gene expression Chao Wang Nov 16, 2005 The Ubiquitin System Ubiquitin (Ub) is a small protein that is composed of 76 amino acids. Found only in eukaryotic organisms and is not found in either eubacteria or archaebacteria. Among eukaryotes, ubiquitin is highly conserved, meaning that the amino acid sequence does not differ much when very different organisms are compared. Ub is a heat-stable protein that folds up into a compact globular structure. It is found throughout the cell and can exist either in free form or as part of a complex with other proteins. In the latter case, Ub is attached (conjugated) to proteins through a covalent bond between the glycine at the C-terminal end of Ub and the side chains of lysine on the proteins. Ub is involved in many cell processes. Regulating the cell cycle, DNA repair, embryogenesis, the regulation of transcription, and apoptosis. Ub is encoded by a family of genes whose translation products are fusion proteins. The Ub genes typically exist in two states : A. The ubiquitin gene (red) can be fused to a ribosomal protein gene (blue) giving rise to a translation product that is a Ub-ribosomal fusion protein; B. Ub genes can exist as a linear repeat, meaning the translation product is comprised of a linear chain of Ub-molecules fused together (a polyubiquitin molecule). Ubiquitin Function Proteins exist as a linear chain of amino acids. This chain can degrade over time as such a reaction is thermodynamically favorable in an aqueous environment. When proteins degrade over time, this is called protein-turnover. It is the balance between a protein's degradation and its synthesis that determines the concentration of that protein inside the cell. Studies of protein turnover rates have shown that some proteins are short-lived while others are long-lived. Long-lived proteins constitute the majority of proteins in the cell. Short-lived proteins are typically key regulatory proteins and abnormal proteins. Ub functions to regulate protein turnover in a cell by closely regulating the degradation of specific proteins. Such a regulatory role is very important. Ub functions in an ATP-dependent fashion. But why is this? We don't need energy to hydrolyze proteins. The reason ATP is required is because machinery is needed to specifically target the proteins that need to be degraded. Ub itself does not degrade proteins. It serves only as a tag that marks proteins for degradation. The degradation itself is carried out by the 26S proteasome. In short, proteins that are to be degraded are first tagged by conjugating them with Ub and these tagged proteins are then recognized and shuttled to the proteasome for degradation. The Ubiquitin-Proteasome Pathway Something else is needed to attach Ub to a protein. Three types of enzymes are needed in most cases. 1. E1 enzymes known as Ub-activating enzymes. These enzymes modify Ub so that it is in a reactive state (making it likely that the C-terminal glycine on Ub will react with the lysine sidechains on the substrate protein). 2. E2 enzymes known as Ub-conjugating enzymes. These enzymes actually catalyze the attachment of Ub to the substrate protein. 3. E3 enzymes known as Ub-ligases. E3's usually function in concert with E2 enzymes, but they are thought to play a role in recognizing the subtrate protein. In yeast, there are many types of E1, E2, and E3 enzymes. For example, 13 different E2 enzymes have been found. While they all carry out the conjugation reaction, they are apparently tailored for specific functions. The general reaction pathway is shown in the figure below. First, Ub is activated by E1 in an ATP-dependent fashion. E2 and E3 then work together to recognize the substrate protein and conjugate Ub to it. Ub can be attached as a monomer or as a previously synthesized chain (as shown). From this point, the ubiquinated protein is shuttled to the proteasome for degradation. What determines if a protein gets tagged by Ub and thus marked for degradation? Let's consider some of signals thus far discovered. 1. The N-degron. In 1986, Varshavsky carried out an elegant set of experiments (Science 234, 179-186) that showed a correlation between the half-life of a protein and its N-terminal residue. This observation gave rise to the so-called N-end rule where one could generally predict the lifespan of a protein by its N-terminal amino acid. 2. Certain amino acid sequences appear to be signals for degradation. One such sequence is known as the PEST sequence because short stretch of about eight amino acids is enriched with proline, glutamic acid, serine, and threonine. 3. Signals may also be buried in the hydrophobic core. This is why partially folded or abnormal, mutant proteins may be prone to degradation. When such proteins exist in their native state, the signals are hidden and the protein is thus long-lived. But in a partially unfolded state, the signals may be seen by the Ub machinery caused the protein to become tagged by Ub. This reaction appears to be hindered by chaperone activity. Ubiquitination lead to protein degradation It is not entirely accurate to think of Ub as a simple tag, as Ub does appear to be involved in degradation. The proteasome is the structure that actually does the degrading. Ubiquiton's degradation role may simply be to decrease the rate of dissociation between proteasomes and interacting substrate proteins. That is, without Ub, proteins may interact with the proteasome, but quickly dissociate. Ub slows down this dissociation. A substrate protein that is conjugated with Ub-chains is thought to interact with a proteasome for a longer period of time, thus increasing the likelihood that the proteasome will degrade it. In fact, Ub could actually function to tether the substrate protein to the proteasome. Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005 Nov 3;438(7064):113-6. California Institute of Technology The ubiquitin–proteasome system (UPS) promotes the destruction of target proteins by attaching to them a ubiquitin chain that is recognized by the 26S proteasome1. The UPS influences most cellular processes, and its targets include transcriptional activators that are primary determinants of gene expression. Emerging evidence indicates that non-proteolytic functions of the UPS might stimulate transcriptional activity. The proteolysis of some transcriptional activators by the UPS can stimulate their function. Authors focused on the role of UPS-dependent proteolysis in the function of inducible transcriptional activators in yeast, and found that inhibition of the proteasome reduced transcription of the targets of the activators Gcn4, Gal4 and Ino2/4. In addition, mutations in SCF-Cdc4, the ubiquitin ligase for Gcn4, or mutations in ubiquitin that prevent degradation, also impaired the transcription of Gcn4 targets. These transcriptional defects were manifested despite the enhanced abundance of Gcn4 on cognate promoters. Proteasome inhibition also decreased the association of RNA polymerase II with Gcn4, Gal4 and Ino2/4 targets, as did mutations in SCF-Cdc4 for Gcn4 targets. Expression of a stable phospho-site mutant of Gcn4 or disruption of the kinases that target Gcn4 for turnover alleviated the sensitivity of Gcn4 activity to defects in the UPS. This mechanism places Gcn4, Gal4 and Ino2/4 into a class of regulatory Factors whose activity is required early in a process but whose subsequent turnover or removal promotes completion of the process or subsequent reaction cycles. We call this phenomenon ‘activation by destruction’ and believe that it might represent a regulatory mechanism for a large class of factors and might be an important determinant of infection and disease. The End