* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Figure 5.x3 James Watson and Francis Crick
History of genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Microevolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA interference wikipedia , lookup
RNA silencing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Frameshift mutation wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Polyadenylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
History of RNA biology wikipedia , lookup
Non-coding RNA wikipedia , lookup
Messenger RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Expanded genetic code wikipedia , lookup
Primary transcript wikipedia , lookup
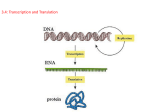
FROM GENE TO PROTEIN Chapter 17 Experiments Leading to Understanding the Link between Genes and Proteins Nirenberg & Matthaei – determining the language of the genetic code. http://bcs.whfreeman.com/thelifewire/content/ chp12/1202002.html Beadle & Tatum – One Gene…One Enzyme hypothesis. http://www.dnalc.org/view/16360-Animation- 16-One-gene-makes-one-protein-.html What is a Gene? Functional definition – a DNA sequence coding for a specific polypeptide chain TEXT PAGE 324 – LEARN THE OVERVIEW…IT WILL HELP YOU MAKE MEANINGFUL UNDERSTANDING OF PROTEIN SYNTHESIS!!! HELPFUL WEBSITES: http://bcs.whfreeman.com/thelifewire/content/chp12/1202001.html http://highered.mcgraw- hill.com/olc/dl/120077/micro06.swf Google – “TRANSCRIPTION & TRANSLATION ANIMATIONS” Overview of Protein Synthesis Genes contain the blueprints for building proteins in cells. RNA is the bridge between DNA and its protein. The “genetic code” is the sequence of bases in DNA that will code for specific amino acids in a growing polypeptide (protein). There are 20 possible amino acids. Types of RNA RNA is a nitrogenous base that consists of a ribose (5 carbon sugar). In most cells, RNA molecules are involved in just one job – protein synthesis! In RNA, the base Uracil is substituted for Thymine in DNA. The RNA nucleic acid is single stranded. The assembly of amino acids into proteins is controlled by RNA There are 3 main types of RNA: 1. 2. 3. Messenger RNA (mRNA) Ribosomal RNA (rRNA) Transfer RNA (tRNA) Protein Synthesis 2 stages: Transcription and Translation Transcription occurs in the nucleus DNA mRNA Making mRNA from a DNA template in the nucleus. Translation occurs in the cytoplasm at a ribosome mRNA protein (using tRNA and rRNA) Translating instructions in mRNA into a growing polypeptide protein chain at the ribosome. Prokaryote vs Eukaryote http://highered.mcgraw-hill.com/olc/dl/120077/bio25.swf Protein Synthesis is about the same in both cases. Here the two steps of Transcription an Translation occur in a bacteria. Prokaryote vs Eukaryote In Eukaryotes one more step occurs. mRNA will be processed before it leaves the nucleus. Segments of mRNA that are not needed will be removed. RNA and Protein Synthesis TYPES OF RNA Messenger RNA (mRNA) – carries information from DNA in the nucleus to the ribosomes where the proteins are assembled. It is a partial copy of ONLY the information needed for that specific job. It is read 3 bases at a time – codon. Ribosomal RNA (rRNA) – found in ribosomes and helps in the attachment of mRNA and in the assembly of proteins. Transfer RNA (tRNA) – transfers the needed amino acids from the cytoplasm to the ribosome so the proteins dictated by the mRNA can be assembled. (The three exposed bases are complementary to the mRNA and are called the anticodon). Types of RNA rRNA Figure 5.28 DNA → RNA → protein: a diagrammatic overview of information flow in a cell Figure 17.3 The triplet code For each gene, one DNA strand functions as a template for transcription – the synthesis of a complementary mRNA molecule. Uracil takes the place of thymine in RNA. During translation, the mRNA is read as a sequence of base triplets (codons). Each codon specifies an amino acid to be added to the growing polypeptide chain. Genes are read in the 3’ to 5’ direction, so mRNA is made in the 5’ to 3’ direction! The Transfer of the Genetic Code The amount of nucleotides that code for an amino acid has to consist of THREE bases. The three amino acid sequence on the mRNA strand is termed a CODON. There are more than one codon for each amino acid due to the 64 combinations possible and only 20 amino acids. Where and What is Happening? The gene will transcribe its self into a mRNA strand each triplet (codon) will code for a specific amino acid. But, How do we know what amino acid? The Codon Chart We can then see what codon will produce what amino acid. You will have to do this. Ex. AGG Ex. CCU Ex. CCT Point A particular triplet of bases in the coding sequence of DNA is AGT. The corresponding codon for the mRNA transcribed is: AGT UCA TCA AGU Either UCA or TCA, depending on wobble in the first base Answer: UCA Point A possible sequence of nucleotides in DNA that would code for the polypeptide sequence pheleu-ile-val would be: 5’ 3’ TTG-CTA’CAG’TAG 3’ AAC-GAC-GUC-AUA 5’ 5’ AUG-CTG-CAG-TAT 3’ 3’ AAA-AAT-ATA-ACA 5’ 3’ AAA-GAA-TAA-CAA 5’ Answer: 3’ AAA-GAA-TAA-CAA 5’ Point Which of the following is correct about a codon? It… Consists of two nucleotides May code for the same amino acid as another Consists of discrete amino acid regions Catalyzes RNA synthesis Is the basic unit of the genetic codon code Answer: may code for the same amino acid as another codon. Section 12-3 Transcription http://bcs.whfreeman.com/thelifewire/content/chp12/1202001.html Adenine (DNA and RNA) Cystosine (DNA and RNA) Guanine(DNA and RNA) Thymine (DNA only) Uracil (RNA only) RNA polymerase DNA RNA During transcription, RNA polymerase uses one strand of DNA as a template to assemble nucleotides into a strand of RNA. Figure 17.6 The stages of transcription: initiation, elongation, and termination (Layer 4) Promoters and Terminators Promoter: sequence in DNA where RNA polymerase attaches and initiates transcription Terminator: sequence that signals the end of transcription Transcription Unit: stretch of DNA that is being copied into mRNA RNA Transcript: stretch of mRNA created using DNA as a template. Point Where is the attachment site for RNA polymerase? Structural gene region Initiation region Promoter region Operator region Regulator region Answer: promoter region Figure 17.7 The initiation of transcription at a eukaryotic promoter http://bcs.whfreeman.com/thelife wire/content/chp14/1402002.html Point Which of the following is LEAST related to the other items? Translation TATA box Transcription Template strand RNA polymerase II Answer: translation RNA Processing Enzymes in the Eukaryotic nucleus must modify pre-mRNA before the genetic message is dispatched to the cytoplasm. This process is called RNA PROCESSING: Both ends of the primary transcript are altered. Addition of 5’ cap and Poly (A) tail Certain interior sections of the molecule are cut out and then the remaining parts are spliced together – called RNA SPLICING. Figure 17.8 RNA processing; addition of the 5 cap and poly(A) tail A modified guanosine triphosphate added to the 5’ end. 50 to 250 adenine nucleotides added to the 3’ end created by cleavage downstream of the termination signal. •Enzymes modify the two ends of a eukaryotic pre-mRNA molecule. •The modified ends help protect the RNA from degradation, and the poly(A) tail may promote the export of mRNA from the nucleus. • When mRNA reaches the cytoplasm, the modified ends, in conjunction with certain cytoplasmic proteins, facilitate ribosome attachment. •The leader and trailer are not translated, nor is the poly(A) tail. Figure 17.9 RNA processing: RNA splicing The gene shown here and its pre-mRNA transcript have three regions, called exons, that consist mostly of coding sequences; exons are separated by noncoding regions, called introns. During RNA processing, the introns are excised and the exons are spliced together. Point What are the coding segments of a stretch of eukaryotic DNA called? Introns Exons Codons Replicons Transposons Answer: exons How is Pre-mRNA Splicing Done? snRNP’s : small nuclear ribonucleoproteins these look for special sequences at ends of introns to know where to cut made up of snRNA (small nuclear RNA) Several different snRNP’s join with additional proteins to form the splicesome – this interacts with the splice sites at intron ends, and cuts them out – then joins the two exons together. http://bcs.whfreeman.com/thelifewire/conte nt/chp14/1402001.html Figure 17.10 The roles of snRNPs and spliceosomes in mRNA splicing 1. Pre-mRNA containing exons and introns combines with small nuclear ribonuceloprotei ns (snRNPs) and other proteins to form a molecular complex call spliceosome. 2. Within the spliceosome, snRNA basepairs with nucleotides at the ends of the intron. 3. The RNA transcript is cut to release the intron, and the exons are spliced together. The spliceosome then comes apart, releasing mRNA, which now contains only exons. Translation Translation is the RNA-directed synthesis of a polypeptide. Generally: is the reading of the codons on the mRNA strand and the sequencing of them into an amino acid sequence – polypeptide. The Players: mRNA: already processed within the nucleus, will be the template for the sequence of amino acids. tRNA: transfers Amino acids from the Cytoplasm to the ribosome. Ribosome: adds amino acids together from the tRNA and in the sequence of the mRNA. Figure 17.15 The anatomy of a functioning ribosome http://highered.mcgrawhill.com/olcweb/cgi/pluginpop.cgi?it=swf::535::535::/sites/dl/free/007243731 6/120077/micro06.swf::Protein%20Synthesis Structure: It is made out of RNA and proteins. The largest type of RNA is ribosomal RNA (rRNA). It has two subunits which will combine to form a ribosome in the cystol. Each ribosome has a binding site for mRNA and three binding sites for tRNA molecules. The P site holds the tRNA carrying the growing polypeptide chain. The A site carries the tRNA with the next amino acid. Discharged tRNAs leave the ribosome at the E site. Point What are ribosomes composed of? Two subunits, each consisting of rRNA only Two subunits, each consisting of several proteins only Both rRNA and protein mRNA, rRNA, and protein mRNA, tRNA, rRNA, and protein Answer: both rRNA and protein Figure 17.13a The structure of transfer RNA (tRNA) tRNA is made in the nucleus. Function: Pick up designated amino acids in the cystol. Deposit the amino acid at the ribosome. Return to the cystol to pick up another amino acid. Structure: It is made out of about 80 nucleotides. It is folded in on its self in a T shape. One important look is called the anti codon. This base pairs with the codon on the mRNA strand. tRNA Amino Acids are placed onto the tRNA by the enzyme aminoacyl tRNA synthase. There are 20 different synthases for the 20 different types of amino acids. Point What is an anticodon a part of? DNA tRNA mRNA A ribosome An activating enzyme Answer: tRNA Translation Translation can be divided into three steps: 1. Initiation 2. Elongation 3. Requires energy (GTP) Requires energy (GTP) Termination Translation http://highered.mcgraw-hill.com/olc/dl/120077/micro06.swf Nucleus Messenger RNA Messenger RNA is transcribed in the nucleus. Phenylalanine tRNA The mRNA then enters the cytoplasm and attaches to a ribosome. Translation begins at AUG, the start codon. Each transfer RNA has an anticodon whose bases are complementary to a codon on the mRNA strand. The ribosome positions the start codon to attract its anticodon, which is part of the tRNA that binds methionine. The ribosome also binds the next codon and its anticodon. Ribosome Go to Section: mRNA Transfer RNA Methionine mRNA Lysine Start codon Translation Continued Translation Initiation • The initiation stage of translation brings together mRNA, a tRNA bearing its first amino acid of the polypeptide, and the two subunits of a ribosome. • All three parts need to be in place for initiation. – Initiator tRNA (Met), Small Ribosomal subunit and Large Ribosomal subunit. • Energy is needed to bind the tRNA to the P site (GTP). Figure 17.17 The initiation of translation A small ribosomal subunit binds to a molecule of mRNA. An initiator tRNA, with the anticodon UAC, base-pairs with the start codon, AUG. This tRNA carries the amino acid methionine (Met). The arrival of a large ribosomal subunit completes the initiation complex. Proteins call initiation factors are required to bring all the translation components together. GTP provides the energy for assembly. The initiator tRNA is in the P site; the A site is available to the tRNA bearing the next amino acid. Translation Elongation – Text 318 Translation elongation occurs in three steps: 1. Codon Recognition 2. Peptide Bond Formation 3. The mRNA codon in the A site of the ribosome forms a H-bond with the anticodon of an incoming molecule of tRNA carrying its appropriate amino acid. This requires energy. (GTP) (A site) The ribosome catalyzes the formation of a peptide bond between the new amino acid and the growing polypeptide. Catalyzed by the ribosome. Translocation The tRNA in the P site and A site are now moved to the E site and P site respectively. The tRNA in the E site will detach and a new codon is open. The ribosome shifts the mRNA by one codon “reading it” This step requires energy. (GTP) Figure 17.18 The elongation cycle of translation Codon Recognition An incoming tRNA binds to the codon in the A site by a H-bond. Peptide Bond Formation The ribosome catalyzes the formation of a peptide bond between the new amino acid and the growing polypeptide. Translocation The tRNA in the A site is translocated to the P site, taking the mRNA along with it. Meanwhile the tRNA in the P site moves to the E site and is released from the ribosome. The ribosome shifts the mRNA by one codon. Figure 17.19 The termination of translation When a ribosome reaches a termination codon on mRNA, the A site of the ribosome accepts a protein called a release factor instead of tRNA. The release factor hydrolyzes the bond between the tRNA and the P site and the last amino acid of the polypeptide chain. The polypeptide in thus freed from the ribosome. The two ribosomal subunits and the other components of the assembly dissociate. Point What is the function of the ribosome in polypeptide synthesis? Answer: Holds mRNA and tRNAs together Catalyzes the addition of amino acids from the tRNAs to the growing polypeptide chain Moves tRNA and mRNA during the translocation process From Polypeptide to Functional Protein During and after its synthesis, a polypeptide chain begins to coil and fold spontaneously, forming a functional protein of specific conformation: A three-dimensional molecule with secondary and tertiary structure. A gene determines the primary structure. The primary structure in turn determines conformation. In many cases, a chaperone protein helps the polypeptide fold correctly. Point Which of the following is NOT directly involved in the process of translation? Ligase tRNA rRNA mRNA Aminoacyl-tRNA Answer: ligase synthase Review – PROTEIN SYNTHESIS Transcription mRNA (in nucleus) In eukaryotes will have RNA processing (in nucleus). DNA Translation mRNA Polypeptide (at ribosome in cytoplasm) Coiling & Folding Three dimensional (in cytoplasm as translation is occurring) Chaperone proteins involved Figure 17.4 The dictionary of the genetic code Section 12-3 Go to Section: The Genetic Code Polyribosomes A single ribosome can make an averagesized polypeptide in less than a minute; Typically, however, a single mRNA is used to make many copies of a polypeptide simultaneously, because a number of ribosomes work on translating the message at the same time. Multiple ribosomes may trail along the same mRNA (in strings called polyribosomes). These help a cell make many copies of a polypeptide very quickly. Mutations Mutations are changes in the genetic material of a cell. Read text pages 322, 323, 325 Point Mutations are chemical changes in just one base pair of a gene. There are two types of point mutations: Base-Pair Base-pair substitution Missense mutation Nonsense mutation Base-Pair Substitutions: Insertions & Deletions: Frameshift mutation Base-Pair Substitutions Base pair substitution is the replacement of one nucleotide and its partner in the complementary DNA strand with another pair of nucleotides. Some substitutions have no effect on the genetic code – it may transform one codon into another that is translated into the same amino acid. CCG mutated to CCA – mRNA codon GGC will become GGU – both code for glycine. Some may switch the amino acid but have little effect on the protein (if amino acid properties are similar). Some may cause a detectable change in a protein: Missense mutations (wrong amino acid coded for & therefore dysfunctional protein produced) Nonsense mutations (amino acid codon changed to a stop codon – no protein produced) Base-Pair Insertions & Deletions Insertions and deletions are additions or losses of nucleotide pairs in a gene. These mutations have disastrous effect on the resulting protein b/c these change the entire triplet code being read – ALL THE WAY DOWN THE mRNA LINE! This occurs when the reading frame (triplet grouping) is altered. Called a frameshift mutation All nucleotides downstream of the mutation will be improperly grouped into codons Unless the frameshift is near the end of a gene, it will produce a protein that is almost certain to be nonfunctional! Gene Mutations: Substitution, Insertion, and Deletion Deletion Substitution Go to Section: Insertion Gene Mutations that only affect ONE point of the code -- often called Point Mutations Figure 17.24 Categories and consequences of point mutations: Base-pair insertion or deletion http://nortonbooks.com/college/biology/ani mations/ch13a08.htm Chromosomal Mutations – affects LARGE portions of the code (entire genes or entire chromosome). Deletion Duplication Inversion Translocation Go to Section: Point A frameshift mutation could result from: A base insertion only A base deletion only A base substitution only Deletion of 3 consecutive bases Either an insertion or a deletion of a base Answer: either an insertion or a deletion of a base So, what is a gene? Functional definition – a DNA sequence coding for a specific polypeptide chain TEXT PAGE 324 – LEARN THE OVERVIEW…IT WILL HELP YOU MAKE MEANINGFUL UNDERSTANDING OF PROTEIN SYNTHESIS!!! HELPFUL WEBSITES: Google – “TRANSCRIPTION & TRANSLATION ANIMATIONS” Point What is the relationship among DNA, a gene, a chromosome, proteins and phenotypes? Answer: A chromosome contains hundreds of genes, which are composed of DNA. The DNA is used as a template for building proteins. Expression of proteins generates certain phenotypes in individuals. The Essay – IT’S A DOOZY! A portion of a specific DNA molecule consists of the following sequence of nucleotide triplets: TAC GAA CTT CGG TCC This DNA sequence codes for the following short polypeptide: methionine - leucine - glutamic acid - proline - arginine a) b) c) d) Describe the steps in the synthesis of this polypeptide. What would be the effect of a deletion or an addition in one of the DNA nucleotides? What would be the effect of a substitution in one of the nucleotides? Cells regulate both protein synthesis and protein activity. Discuss TWO specific mechanisms of protein regulation in eukaryotic cells.