* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download AP Biology
Gene expression profiling wikipedia , lookup
DNA vaccination wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene desert wikipedia , lookup
Transposable element wikipedia , lookup
Genetic engineering wikipedia , lookup
Molecular cloning wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Gene nomenclature wikipedia , lookup
Koinophilia wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genome evolution wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genetic code wikipedia , lookup
Pathogenomics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Human genome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA barcoding wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Computational phylogenetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microsatellite wikipedia , lookup
Sequence alignment wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Metagenomics wikipedia , lookup
Genome editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
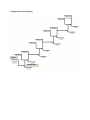
AP Biology Name_________________________________________ AP Lab Three: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST In the 1990’s when scientists began to compile a list of genes and DNA sequences in the human genome it became abundantly clear that we were eventually going to need a place to put all of these sequences. One of the systems developed was BLAST, or Basic Local Alignment Search Tool. The BLAST computer program uses complicated algorithms to search through over 21 million DNA, RNA and protein sequences of a maximum 10,000 base pairs. With a few clicks of a mouse students and scientists alike can compare known and unknown DNA sequences, establish common relationships between organisms, and look for similar protein structures in different organisms. All in a matter of seconds. This lab is an activity introducing you to using this amazing computer program. The first part of the activity is required. All other sections are extra credit. Activity #1: Constructing a cladogram from known traits (Required) Cladograms are visual trees that quickly show an evolutionary relationship between organisms based on a specific trait. Cladograms typically show established, linked relationships, but they may also be simply based on guesses. As a cladogram line increases up, the relationships between organisms become closer. In the above cladogram, the closest relationship is that of the crocodiles and birds while the least-related organisms are the sharks and the birds. Amphibians are more closely related to primates than to rodents and rabbits, and sharks emerged earlier in earth’s history than ray-finned fish did. Cladograms also provide a hypothesis for what evolutionary trait separated different groups of organisms. In the above example, all organisms in the cladogram have vertebrae, but the sharks do not have a bony skeleton. Primates, rodents and rabbits, crocodiles and birds all have eggs, but only crocodiles and birds have eggs with shells. Part A: Below are 8 species of animals, organized alphabetically. Use the pictures or research different traits that these organisms have. Then, hypothesize which organisms are the oldest/youngest on the planet and what traits arose first in history. Hypothesize which trait separated each species and which species are the oldest/newest. Show your hypotheses by filling in Cladogram #1, placing the species and the traits in their appropriate locations. BARNACLE CRAB LAMPREY SEA SLUG SEA URCHIN SKATE SQUID STARFISH Part B: Below is a table of traits and whether or not each of the plant organisms has that particular trait. Use these traits to determine which organisms are the oldest/youngest on the planet and what traits arose first in history. Write your answers on the second cladogram (Cladogram #2). Conifer Cycad Fern Horsetail Liverwort Moss Peach Rose Enclosed Embryo Flowers Fruits Leaves Seeds Stomata Vascular Tissue Wood YES YES YES YES YES YES YES YES NO NO NO NO NO NO YES YES NO NO NO NO NO NO YES NO YES YES YES NO NO NO YES YES YES YES NO NO NO NO YES YES YES YES YES YES NO YES YES YES YES YES YES YES NO NO YES YES YES NO NO NO NO NO YES YES Activity #2: Constructing a cladogram from known and unknown DNA sequence (Required) Using BLAST, a scientist compared a human copy of GAPDH (glyceraldehyde-3-phosphate dehydrogenase), an important reaction in glycolysis. The data table below shows the similarities between the DNA sequences and the protein sequences of human GAPDH and the GAPDH of other organisms. Species Chimpanzee (Pan troglodytes) Dog (Canis lupus familiaris) Fruit Fly (Drosophila melanogaster) Roundworm (Caenorhabditis elegans) Gene Percentage Similarity Protein Percentage Similarity 99.6% 91.3% 72.4% 68.2% 100% 95.2% 76.7% 74.3% 1. On a separate sheet of paper, draw a phylogenetic cladogram of all five species (including humans) according to the gene percentage similarity. 2. On the paper, answer the following question: Why is the percentage of similarity in the gene always lower than the percentage of similarity in the protein for each of the species? Activity #3: Constructing a cladogram using BLAST (Required) A team of scientists has uncovered a fossil specimen near Liaoning Province, China. Little is known about the fossil. It appears to be a new species. Upon careful examination of the fossil, small amounts of soft tissue have been discovered. Normally, soft tissue does not survive fossilization; however, rare situations of such preservation do occur. Scientists were able to extract DNA nucleotides from the tissue and use the information to sequence several genes. Your task is to use BLAST to analyze these genes and determine the most likely placement of the fossil species on Figure 4. 1. Obtain a card with one of the four gene sequences written down. This will be the fossil you research and build your cladogram from. 2. At the top of the page, click on the tab marked “Saved Strategies.” 3. Under “Upload Search Strategy,” click on “Browse”. Follow the instructions on the board or website to locate Marshall’s shared drive. Click on the gene sequence that matches your assigned gene. 4. The screen will appear pre-loaded according to the necessary parameters you will use. Scroll down and click the button on the bottom left marked “BLAST.” DO NOT CHANGE ANY PARAMETERS. 5. When the results page appears, scroll down to the section titled “Sequences producing significant alignments.” This is a list of all gene sequences that show similarities to the fossil sequence, the most similar being on top. 6. Choose any four of the top 20 sequences. Each sequence includes information about the species it was taken from, including the scientific name. Research these scientific names and construct a table to display the following data: a. Common names b. Similar characteristics between the four organisms c. Similarity to the unknown fossil based on the “Max Ident” number. d. Species identification of the unknown fossil according to BLAST. 7. Construct a cladogram showing the evolutionary relationships of your species. 8. After you have a completed cladogram, show Marshall and he will show you the actual cladogram to compare to yours. a. What similarities did your cladogram and the actual one have? What differences? b. If you were constructing a cladogram based on physical relationships rather than genetic relationships, what similarities would it share? What differences? Activities #4-6: Extra Credit Activities 1. Activity #1: Comparing sequences of DNA a. Run a BLAST on gene sequence #1. Click on the top sequence in the “Sequences producing significant alignments” list. b. Record the first 10 nucleotides in the sequence. Scroll down to the end of the sequence. Record the last 10 nucleotides in the sequence. c. Repeat for the other three gene sequences. d. Questions i. What similarities or patterns do you see in the initial or terminal sequences for the gene? ii. What is a possible reason for why genes have this pattern at the ends of their sequences? Explain your reasoning. 2. Activity #2: Variety of DNA sequences a. Run a BLAST on any gene sequence of your choosing. Click on the top sequence in the “Sequences producing significant alignments” list. b. In your lab book, write down any random 4 DNA-nucleotide sequence. Also write any random 5 DNA, 6 DNA, and 7 DNA nucleotide sequence. c. For the 4, 5, 6, and 7 nucleotide sequence, hypothesize how often this sequence will appear in your gene. Record in your lab book. d. While your gene sequence is on your computer screen, type “CTRL+F” to pull up the “search” function. Type your 4-nucleotide sequence into the screen. Count the number of times your sequence ACTUALLY appears in your gene. Record. e. Repeat for your 5, 6, and 7-nucleotide sequences. f. Questions i. Build a table to organize the information you’ve recorded. ii. What does this lab teach you about the possible number of different gene sequences based on only four different nucleotide options (A,C,T, and G)? Explain your reasoning. iii. You want to cut up your gene for an electrophoresis experiment. Assuming you use a restriction enzyme that codes for your 5-DNA nucleotide sequence, how many fragments would be in your gel electrophoresis stain? 3. Activity #3: Protein sequencing a. Run a BLAST on gene sequence #1. Click on the top sequence in the “Sequences producing significant alignments” list. b. At the top of the sequence, locate the label for “Length.” This tells you the number of DNA nucleotides in the sequence. c. Predict how many amino acids this sequence will code for during translation. Also, predict what the first amino acid will be. d. On the right side of the page, click the link marked “gene”. e. On the right side of this page, click the link marked “RefSeq Proteins” under the “Related Information” section. f. At the top of the page is the label for how many amino acids (a.a.) are in the protein. g. Scroll down to view the amino acid sequence. Each amino acid is represented by one letter. h. Questions i. What is the actual first amino acid in the sequence? How does this compare to your guess? ii. Compare the actual number of amino acids to your prediction. Were they the same? If not, what could account for the difference? To earn credit for AP Lab 3, you will need to submit the following items 1. 2. 3. 4. 5. 6. 7. Cladogram #1 Cladogram #2 Activity #2: phylogenetic cladogram, answer to question #2 attached BLAST data table BLAST cladogram Answers to BLAST cladogram questions. If you perform any of the extra credit activities, include their necessary requirements as well Cladogram #1: Animal Phylogeny PLANTS HETEROTROPH Cladogram #2: Plant Phylogeny ALGAE CLOSED EMBRYO