* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Investigation #3
Genomic imprinting wikipedia , lookup
Pathogenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene therapy wikipedia , lookup
Metagenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Transitional fossil wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome (book) wikipedia , lookup
Gene desert wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Point mutation wikipedia , lookup
Genome evolution wikipedia , lookup
The Selfish Gene wikipedia , lookup
Smith–Waterman algorithm wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Sequence alignment wikipedia , lookup
Gene expression profiling wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Multiple sequence alignment wikipedia , lookup
Helitron (biology) wikipedia , lookup
Designer baby wikipedia , lookup
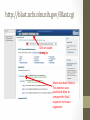
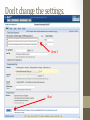
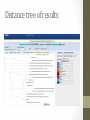
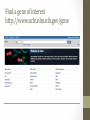
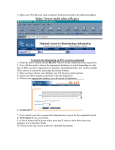
Investigation #3 Hmm…Make some observations. Go to http://www.amnh.org/learn/pd/dinos/gallery. html for a high resolution photo. This fossil was found near Liaoning Province China. Where does this organism “belong” in the cladogram on the next page? DNA sequence of several genes was obtained. Use this to place the organism. Fossil Cladogram Crocodilians Palatal valve Insects Exposed mouthparts Birds Great Apes Opposable feather thumbs Crustaceans Two-parted limbs Rodents Two specialized incisors fur vertebrae heterotroph From page S45 AP Biology Investigative Labs from the College Board. Download 3 of 4 files • http://blogging4biology.edublogs.org/2010/08/28/collegeboard-lab-files • Don’t open. http://blast.ncbi.nlm.nih.gov/Blast.cgi Click on saved strategies Many data base choices. This exercise uses nucleotide blast to compare the fossil sequence to known organisms. Browse for one of the saved files Don’t change the settings. Gene 1 Blast Analysis Gene entered Red bars compare alignment with the query. Number of results Scroll down. Listed in order of similarity This score refers to gaps and substitutions. Higher=more similar alignment E value is the likelihood a match occurred by chance. Lower the number the better the match. Refer to other data bases Distance tree of results Repeat with other genes • Think about your hypothesis in response to the question: Where does the fossil fit on your cladogram? • Remember: • The higher the score, the closer the alignment. • The lower the e value, the closer the alignment. • Sequences with e values less than 1e-04 (1 x 10-4) can be though of as related • See page S48 for additional questions and some thoughts on designing your own investigation. From the College Board AP Biology Investigative Labs. Find a gene of interest http://www.ncbi.nlm.nih.gov/gene