* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Powerpoint file
Epigenetics in stem-cell differentiation wikipedia , lookup
Public health genomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
X-inactivation wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Oncogenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
RNA interference wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Essential gene wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Messenger RNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Primary transcript wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Minimal genome wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Designer baby wikipedia , lookup
Ridge (biology) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
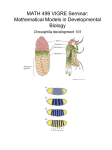
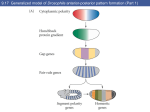
Chapter 18 Gene Regulation During Development 7 and 9 November, 2004 • • • • • • • • • • Overview Differences in cell type are fundamentally differences in gene expression. These expression differences are often monitored using microarray hybridization. Differential gene expression is initiated by asymmetrical mRNA distribution, cellcell contact, or by diffusible signals. Gradients of signaling molecules can give location information to cells. Asymmetrically distributed mRNAs use the cytoskeleton for localization. Drosophila embryogenesis is a well-understood system of animal development. A morphogen gradient controls dorsal-ventral orientation in Drosophila. Asymmetrically distributed mRNAs control anterior-posterior orientation and ultimately segmentation. Segmentation also depends on the interplay of regulators at the control regions of genes. Bioinformatics tools are useful in identifying genes that are developmentally regulated. Signaling Differentiation Transducing the Signal Regualtion in a Morphogen Gradient Regulation by Asymmetrically Distributed mRNA Regulation by Asymmetrically Distributed mRNA Polar Cytoskeletal Elements Regulation by Cell-cell contact in B. subtilis Regulation by Cell-cell contact in Animal Development Regulation by Cell-cell contact in Animal Development Morphogen Gradient Drosophila Development Spätzle-Toll create a gradient of Dorsal in nuclei. Transcriptional response to the Dorsal Gradient Snail keeps rhomboid off in cells of the mesoderm. Polar Distribution of bicoid and oksar in Drosophila Embryos Bicoid regulates orthodenticle and zygotic hunchback. Maternal hunchback mRNA is evenly distributed, but translationally repressed by the posteriorly concentrated nanos. The hunchback gradient regulates gap genes as a transcriptional repressor. Gap genes direct the expression of pair-rule genes. Gap genes direct the expression of pair-rule genes. Bioinformatics and the identification of enhancers. Competition and Quecnching in the eve enhancer Repression of eve-3 and eve-4 enhancers by hunchback and knirps. Enhancer autonomy requires short-range repression. Title Title