* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download DNA to Protein
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Biochemistry wikipedia , lookup
Bottromycin wikipedia , lookup
List of types of proteins wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Expanded genetic code wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA interference wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic code wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Polyadenylation wikipedia , lookup
Messenger RNA wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene expression wikipedia , lookup
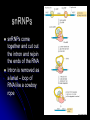
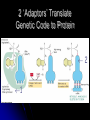
Chapter 7 From DNA to Protein DNA to Protein DNA acts as a “manager” in the process of making proteins DNA is the template or starting sequence that is copied into RNA that is then used to make the protein Central Dogma One gene – one protein Central Dogma This is the same for bacteria to humans DNA is the genetic instruction or gene DNA RNA is called Transcription RNA chain is called a transcript RNA Protein is called Translation Expression of Genes Some genes are transcribed in large quantities because we need large amount of this protein Some genes are transcribed in small quantities because we need only a small amount of this protein Transcription Copy the gene of interest into RNA which is made up of nucleotides linked by phosphodiester bonds – like DNA RNA differs from DNA Ribose is the sugar rather than deoxyribose – ribonucleotides U instead of T; A, G and C the same Single stranded Can fold into a variety of shapes that allows RNA to have structural and catalytic functions RNA Differences RNA Differences Transcription Similarities to DNA replication Open and unwind a portion of the DNA 1 strand of the DNA acts as a template Complementary base-pairing with DNA Differences RNA strand does not stay paired with DNA DNA re-coils and RNA is single stranded RNA is shorter than DNA RNA is several 1000 bp or shorter whereas DNA is 250 million bp long Template to Transcripts The RNA transcript is identical to the NONtemplate strand with the exception of the T’s becoming U’s Catalyzes the formation of the phosphodiester bonds between the nucleotides (sugar to phosphate) Uncoils the DNA, adds the nucleotide one at a time in the 5’ to 3’ fashion Uses the energy trapped in the nucleotides themselves to form the new bonds RNA Polymerase RNA Elongation Reads template 3’ to 5’ Adds nucleotides 5’ to 3’ (5’ phosphate to 3’ hydroxyl) Synthesis is the same as the leading strand of DNA RNA Polymerase RNA is released so we can make many copies of the gene, usually before the first one is done Can have multiple RNA polymerase molecules on a gene at a time Differences in DNA and RNA Polymerases RNA polymerase adds ribonucleotides not deoxynucleotides RNA polymerase does not have the ability to proofread what they transcribe RNA polymerase can work without a primer RNA will have an error 1 in every 10,000 nucleotides (DNA is 1 in 10,000,000 nucleotides) Types of RNA messenger RNA (mRNA) – codes for proteins ribosomal RNA (rRNA) – forms the core of the ribosomes, machinery for making proteins transfer RNA (tRNA) – carries the amino acid for the growing protein chain DNA Transcription in Bacteria RNA polymerase must know where the start of a gene is in order to copy it RNA polymerase has weak interactions with the DNA unless it encounters a promoter A promoter is a specific sequence of nucleotides that indicate the start site for RNA synthesis RNA Synthesis RNA pol opens the DNA double helix and creates the template RNA pol moves nt by nt, unwinds the DNA as it goes Will stop when it encounters a STOP codon, RNA pol leaves, releasing the RNA strand Sigma () Factor Part of the bacterial RNA polymerase that helps it recognize the promoter Released after about 10 nucleotides of RNA are linked together Rejoins with a released RNA polymerase to look for a new promoter Start and Stop Sequences DNA Transcribed The strand of DNA transcribed is dependent on which strand the promoter is on Once RNA polymerase is bound to promoter, no option but to transcribe the appropriate DNA strand Genes may be adjacent to one another or on opposite strands Eukaryotic Transcription Transcription occurs in the nucleus in eukaryotes, nucleoid in bacteria Translation occurs on ribosomes in the cytoplasm mRNA is transported out of nucleus through the nuclear pores RNA Processing Eukaryotic cells process the RNA in the nucleus before it is moved to the cytoplasm for protein synthesis The RNA that is the direct copy of the DNA is the primary transcript 2 methods used to process primary transcripts to increase the stability of mRNA being exported to the cytoplasm RNA capping Polyadenylation RNA Processing RNA capping happens at the 5’ end of the RNA, usually adds a methylgaunosine shortly after RNA polymerase makes the 5’ end of the primary transcript Polyadenylation modifies the 3’ end of the primary transcript by the addition of a string of A’s Coding and Non-coding Sequences In bacteria, the RNA made is translated to a protein In eukaryotic cells, the primary transcript is made of coding sequences called exons and non-coding sequences called introns It is the exons that make up the mRNA that gets translated to a protein RNA Splicing Responsible for the removal of the introns to create the mRNA Introns contain sequences that act as cues for their removal Carried out by small nuclear riboprotein particles (snRNPs) snRNPs snRNPs come together and cut out the intron and rejoin the ends of the RNA Intron is removed as a lariat – loop of RNA like a cowboy rope Benefits of Splicing Allows for genetic recombination Link exons from different genes together to create a new mRNA Also allows for 1 primary transcript to encode for multiple proteins by rearrangement of the exons Summary RNA to Protein Translation is the process of turning mRNA into protein Translate from one “language” (mRNA nucleotides) to a second “language” (amino acids) Genetic code – nucleotide sequence that is translated to amino acids of the protein Degenerate DNA Code Nucleotides read 3 at a time meaning that there are 64 combinations for a codon (set of 3 nucleotides) Only 20 amino acids More than 1 codon per AA – degenerate code with the exception of Met and Trp (least abundant AAs in proteins) Reading Frames Translation can occur in 1 of 3 possible reading frames, dependent on where decoding starts in the mRNA Transfer RNA Molecules Translation requires an adaptor molecule that recognizes the codon on mRNA and at a distant site carries the appropriate amino acid Intra-strand base pairing allows for this characteristic shape Anticodon is opposite from where the amino acid is attached Wobble Base Pairing Due to degenerate code for amino acids some tRNA can recognize several codons because the 3rd spot can wobble or be mismatched Allows for there only being 31 tRNA for the 61 codons Attachment of AA to tRNA Aminoacyl-tRNA synthase is the enzyme responsible for linking the amino acid to the tRNA A specific enzyme for each amino acid and not for the tRNA 2 ‘Adaptors’ Translate Genetic Code to Protein 2 1 Ribosomes Complex machinery that controls protein synthesis 2 subunits 1 large – catalyzes the peptide bond formation 1 small – binds mRNA and tRNA Contains protein and RNA rRNA central to the catalytic activity Folded structure is highly conserved Protein has less homology and may not be as important Ribosome Structures May be free in cytoplasm or attached to the ER Subunits made in the nucleus in the nucleolus and transported to the cytoplasm Ribosomal Subunits 1 large subunit – catalyzes the formation of the peptide bond 1 small subunit – matches the tRNA to the mRNA Moves along the mRNA adding amino acids to growing protein chain Ribosomal Movement E-site 4 binding sites mRNA binding site Peptidyl-tRNA binding site (P-site) Aminoacyl-tRNA binding site (A-site) Holds tRNA attached to growing end of the peptide Holds the incoming AA Exit site (E-site) 3 Step Elongation Phase Elongation is a cycle of events Step 1 – aminoacyl-tRNA comes into empty A-site next to the occupied P-site; pairs with the codon Step 2 – C’ end of peptide chain uncouples from tRNA in P-site and links to AA in A-site Peptidyl transferase responsible for bond formation Each AA added carries the energy for the addition of the next AA Step 3 – peptidyl-tRNA moves to the Psite; requires hydrolysis of GTP tRNA released back to the cytoplasmic pool Initiation Process Determines whether mRNA is synthesized and sets the reading frame that is used to make the protein Initiation process brings the ribosomal subunits together at the site where the peptide should begin Initiator tRNA brings in Met Initiator tRNA is different than the tRNA that adds other Met Ribosomal Assembly Initiation Phase Initiation factors (IFs) catalyze the steps – not well defined Step 1 – small ribosomal subunit with the IF finds the start codon –AUG Moves 5’ to 3’ on mRNA Initiator tRNA brings in the 1st AA which is always Met and then can bind the mRNA Step 2 – IF leaves and then large subunit can bind – protein synthesis continues Met is at the start of every protein until post-translational modification takes place Eukaryotic vs Procaryotic Procaryotic No CAP; have specific ribosome binding site upstream of AUG Polycistronic – multiple proteins from same mRNA Eucaryotic Monocistronic – one polypeptide per mRNA Protein Release Protein released when a STOP codon is encountered UAG, UAA, UGA (must know these sequences!) Cytoplasmic release factors bind to the stop codon that gets to the A-site; alters the peptidyl transferase and adds H2O instead of an AA Protein released and the ribosome breaks into the 2 subunits to move on to another mRNA As the ribosome moves down the mRNA, it allows for the addition of another ribosome and the start of another protein Each mRNA has multiple ribosomes attached, polyribosome or polysome Polyribosomes Regulation of Protein Synthesis Lifespan of proteins vary, need method to remove old or damaged proteins Enzymes that degrade proteins are called proteases – process is called proteolysis In the cytosol there are large complexes of proteolytic enzymes that remove damaged proteins Ubiquitin, small protein, is added as a tag for disposal of protein Protein Synthesis Protein synthesis takes the most energy input of all the biosynthetic pathways 4 high-energy bonds required for each AA addition 2 in charging the tRNA (adding AA) 2 in ribosomal activities (step 1 and step 3 of elongation phase) Summary Ribozyme A RNA molecule can fold due to its single stranded nature and in folding can cause the cleavage of other RNA molecules A RNA molecule that functions like an enzyme hence ribozyme name