* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt - Chair of Computational Biology
Gene expression programming wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Public health genomics wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Primary transcript wikipedia , lookup
History of genetic engineering wikipedia , lookup
Transposable element wikipedia , lookup
X-inactivation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Human genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Point mutation wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Minimal genome wikipedia , lookup
Designer baby wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Pathogenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Genome editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
V3 regulation of imprinted genes
Review of lecture V2 ...
What does the differential methylation
of CpG islands mean?
- What do the two models describe?
- How did the authors arrive at
the two models?
- How could one distinguish between
these two models?
Biological Sequence Analysis
SS 2008
lecture 3
1
Outline
•
what is genomic imprinting?
•
networks of imprinted genes
•
imprinting mechanisms
•
protein-DNA interaction hypotheses
•
detecting motifs in DNA sequences
– evolutionary conserved regions
– protein binding sites
– gene regulation modules
– "imprinting motifs"
– ...
•
Alu sequences
•
KCNQ1
Biological Sequence Analysis
SS 2008
lecture 3
2
Genomic Imprinting
Igf2
H19
Igf2
Igf2
•
monoallelic expression of a
gene depending on its
parental origin
•
in mammals about 70 known
imprinted genes (human,
mouse), also found in insects
and in flowering plants
•
estimated: 1 - 2% of all genes
= 300 - 600
•
imprinted genes are often
organized in clusters with
"imprinting centers"
H19
H19
paternal gene copy
maternal gene copy
Igf2: coding insulin-like growth factor protein
H19: untranslated RNA
Biological Sequence Analysis
SS 2008
lecture 3
3
Imprinted genes
Imprinted genes of the mouse are
distributed unevenly throughout
the genome.
About half of the known ones are
located on Chromosome 7,
clustered into at least five distinct
imprinted domains.
red: maternally expressed genes
blue: paternally expressed genes
PLOS Genet. 2, e147
(2006)
Biological Sequence Analysis
SS 2008
lecture 3
4
Imprinting Mechanisms
•
methylation of Cytosine in CpG: differentially methylated regions (DMRs)
• altered chromatin structure
NH2
• binding of proteins (transcription factors, silencers)
depending on methylation status
•
NH2
N
O
N
O
N
H
H
setting the imprint
– hypothesis: male specific and female germ line specific proteins
recognize different patterns and set different imprints in sperm and egg
– how these imprint markers might find their targets:
• tandem repeats
– sequence not (well) conserved – like many DMRs –
– are enriched in the CpG islands of imprinted genes
– special DNA structure
• sequence patterns (germ line specific protein/transcription factor
binding sites): evolutionary conserved
AGAACCGCGGCGAGAGGCC
Biological Sequence Analysis
SS 2008
lecture 3
CH3
N
AGAACCGCGCCGAAGAACC
ACAACCGCGCCGAAGAACC
AGAACCGCGCCGAAAAGCC
5
Regulatory models at imprinted loci
(A) The enhancer–blocker model (also known
as the boundary model) is well studied at the
Igf2/H19 locus and consists of an imprinting
control region (ICR) located between a pair of
reciprocally expressed genes that controls
access to shared enhancer elements.
On the paternal allele, the differentially
methylated domain (DMD) acquires
methylation (black circles) during
spermatogenesis, which leads to repression of
the H19 promoter. The hypomethylated
maternal DMD acts as an insulator element,
mediated through binding sites for the
methylation-sensitive boundary factor CTCF
(shaded ellipse). When CTCF is bound, Igf2
promoter access to the enhancers (E) distal to
H19 is blocked.
Blue boxes : paternally expressed
alleles,
red boxes : maternally expressed
alleles,
black boxes : silenced alleles, grey
boxes : nonimprinted genes.
Arrows on boxes indicate
transcriptional orientation.
PLOS Genet. 2, e147
(2006)
Biological Sequence Analysis
SS 2008
lecture 3
6
Protein Interactions and Chromatin Loops
•
•
reading the imprint:
candidate "imprinting
transcription factors"
CTCF, YY1
chromatin loop model
– DMRs interact via
proteins
– mediates
interaction with
the enhancers
H19
p
m
Igf2
p
m
Murrell et al. (2004) Nature Genet. 36: 889
• maternal chromosome: DMR1 and DMR unmethylated, CTFC bound H19 is
expressed (interaction with the enhancers), Igf2 is silenced
• paternal chromosome: DMR and DMR2 methylated, no CTCF binding Igf2 in
contact with enhancers, active; H19 silenced
Biological Sequence Analysis
SS 2008
lecture 3
7
Regulatory models at imprinted loci
(B) At the Igf2r locus on Chromosome
17, the paternally expressed, noncoding
RNA Air acts to induce bidirectional cismediated silencing (black curved lines)
on neighbouring protein-coding genes
(maternally expressed Igf2r, Slc22a3,
and Slc22a2).
The grey ellipses are the intronic imprint
control elements that are maternally
methylated (black circles) and contain
the promoter of the Air RNA.
PLOS Genet. 2, e147
(2006)
Biological Sequence Analysis
SS 2008
lecture 3
8
Regulatory models at imprinted loci
(C) At microimprinted domains,
oocyte-derived methylation in the
promoter region of a proteincoding gene is likely to be the
primary epigenetic mark leading
to monoallelic silencing. With the
exception of the U2af1-rs1 locus,
the multiexonic genes within
which the paternally expressed
transcripts are embedded, escape
imprinting. The paternally
expressed Nap1l5 is situated
within intron 22 of Herc3, which is
expressed from both alleles.
PLOS Genet. 2, e147
(2006)
Biological Sequence Analysis
SS 2008
lecture 3
9
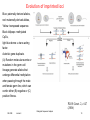
Evolution of imprinted loci
Blue: paternally derived alleles,
red: maternally derived alleles,
Yellow: transposed sequence.
Black lollipops: methylated
CpGs,
light blue dome: a trans-acting
factor.
Asterisk: gene duplicate.
(A) Random molecular events or
mutations in the germ-cell
lineage generate alleles that
undergo differential methylation
when passing through the male
and female germ line, which can
confer either (B) negative or (C)
positive fitness.
PLOS Genet. 2, e147
(2006)
Biological Sequence Analysis
SS 2008
lecture 3
10
Functions of Imprinted Genes
•
imprinting disorders generally cause diseases
– over- or underexpression of the corresponding gene products
•
control cell proliferation
– growth factors
– tumor suppressors
– embryonic development ("giant baby")
•
important for brain development
– (adult) behavior
•
imprinted genes are often transcription factors
– regulation of other genes
Biological Sequence Analysis
SS 2008
lecture 3
11
Are the imprinted genes alone?
Protein-Protein Interactions of Imprinted Genes
•
Source: Unified Human Interactome (http://mdc-berlin.de/unihi)
Biological Sequence Analysis
SS 2008
lecture 3
12
Are the imprinted genes alone?
Coexpression Network of Imprinted Genes
http://symatlas.gnf.org
Varrault et al. (2006) Dev. Cell 11: 711
Zac1 (Plagl1) is a transcription factor -> also regulatory networks!
Arima et al. (2005) NAR 33: 2650
Biological Sequence Analysis
SS 2008
lecture 3
13
Imprinted genes and repetitive elements
Imprinted genes show depletion of short interspersed transposable elements
(SINEs)
and an enrichment of long interspersed nuclear element 1 (LINE-1) repeats.
Biological Sequence Analysis
SS 2008
lecture 3
14
Alu sequence
An Alu sequence is a short stretch 300 bp of DNA originally characterized by the
action of the Alu restriction endonuclease that was isolated from Arthrobacter
luteus. They are therefore classified as short interspersed nuclear elements
(SINEs) and are the most abundant mobile elements in the human genome.
There are over one million Alu sequences of different kinds interspersed
throughout the human and other primate genomes, and probably make up about
10% of the whole genome. Less than 0.5% are polymorphic.
Alu sequences are derived from the small cytoplasmic 7SL RNA, a component of
the signal recognition particle.
The recognition sequence of the Alu endonuclease is 5' AG/CT 3.
Most human Alu sequence insertions can be found in the corresponding positions
in the genomes of other primates. About 7,000 Alu insertions are unique to
humans.
www.wikipedia.org
Biological Sequence Analysis
SS 2008
lecture 3
15
variability of Alu sequences
Nat. Rev. Gen. 3,
370 (2002)
Biological Sequence Analysis
SS 2008
lecture 3
16
insertion of Alu sequence
Alu elements are thought to
„borrow“ factors such as a
functional reverse transcriptase
from nearby LINE elements.
Nat. Rev. Gen. 3,
370 (2002)
Biological Sequence Analysis
SS 2008
lecture 3
17
history of Alu sequences
Most Alu repeats duplicated
ca. 40 Mya.
At that time ca. 1 new Alu
insertion every primate birth.
Currently, ca. 1 Alu insertion
every 200 births.
Possible reasons for decline:
- altered transcription or
reverse transcription activity
- decreased availability of
available insertion sites.
Nat. Rev. Gen. 3,
370 (2002)
Biological Sequence Analysis
SS 2008
lecture 3
18
Spread of an Alu insertion
Nat. Rev. Gen. 3,
370 (2002)
Biological Sequence Analysis
SS 2008
lecture 3
19
Example for imprinted gene: KCNQ1 – KvLQT1
• KvLQT1 is a potassium channel protein coded for by the gene KCNQ1.
KvLQT1 is present in the cell membranes of cardiac muscle tissue and in inner
ear neurons among other tissues. In the cardiac cells, KvLQT1 mediates the IKs
(or slow delayed rectifying K+) current that contributes to the repolarization of the
cell, terminating the cardiac action potential and thereby the heart's contraction.
• Mutations in the gene can lead to a defective protein and several forms of
inherited arrhythmias as Long QT syndrome, Short QT syndrome, and Familial
Atrial Fibrillation.
• The gene product can form heteromultimers with two other potassium channel
proteins, KCNE1 and KCNE3. The gene is located in a region of chromosome 11
that contains a large number of contiguous genes that are abnormally imprinted
in cancer and the Beckwith-Wiedemann syndrome. Two alternative transcripts
encoding distinct isoforms have been described
www.wikipedia.org
Biological Sequence Analysis
SS 2008
lecture 3
20
2D structure of KCNQ1
comment: S1 – S6 are six transmembrane helices
the P-loop between S5 and S6 enters into the membrane and forms the
selectivity pore.
Smith et al. Biochemistry (2007) 46, 14141
Biological Sequence Analysis
SS 2008
lecture 3
21
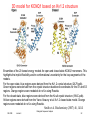
3D model for KCNQ1 based on Kv1.2 structure
Ensembles of the 20 lowest energy models for open and closed state KCNQ1 monomers. This
highlights the implicit flexibility and/or conformational uncertainty for the loop segments of the
models.
For the open state, blue regions were derived from the Kv1.2 crystal structure (2A79.pdb).
Green regions were derived from the crystal structure backbone coordinates for the S1 and S3
regions. Orange regions were modeled de noVo using Rosetta.
For the closed state, blue regions were derived from the KcsA crystal structure (1K4C.pdb).
Yellow regions were derived from the Yarov-Yaravoy et al. Kv1.2 closed state model. Orange
regions were modeled de noVo using Rosetta.
Smith et al. Biochemistry (2007) 46, 14141
Biological Sequence Analysis
SS 2008
lecture 3
22
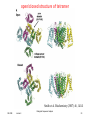
open/closed structure of tetramer
Smith et al. Biochemistry (2007) 46, 14141
Biological Sequence Analysis
SS 2008
lecture 3
23
KCNQ1 – position of disease associated mutations
Smith et al. Biochemistry (2007) 46, 14141
Biological Sequence Analysis
SS 2008
lecture 3
24
CpG islands
„rich“ in CG pairs
more precisely: not so „poor“ in CG pairs as the rest of the genome
recall that Cs in CpGs are often deamidated and converted into Ts.
Why are CpG islands often found at the promoter region?
Because this region is under high selective pressure.
Biological Sequence Analysis
SS 2008
lecture 3
25
•
What are Tandem repeats?
•
How does one find CpG islands?
•
What are Gardiner-Frommer and Takai-Jones parameters?
•
Why do we need t-tests?
•
What are the findings of this paper?
Biological Sequence Analysis
SS 2008
lecture 3
26