* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Exercises Biological databases PART
Whole genome sequencing wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Human genetic variation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Protein moonlighting wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Transposable element wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Copy-number variation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genomic library wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Point mutation wikipedia , lookup
Gene desert wikipedia , lookup
Metagenomics wikipedia , lookup
Human genome wikipedia , lookup
Human Genome Project wikipedia , lookup
Microevolution wikipedia , lookup
Pathogenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Genome editing wikipedia , lookup
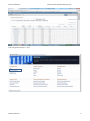
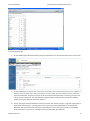
Exercise databases Bioinformatics (updated 2015 january) EXERCISES BIOLOGICAL DATABASES PART Exercises Biological databases PART .......................................................................................................................... 1 Discovering genome projects in NCBI ............................................................................................................................. 1 View the genome sequence initiatives (Go to Bioproject) ......................................................................................... 1 Get a look at the large initiatives: ............................................................................................................................... 6 Using the Entrez search engine to discover distinct databases at NCBI ......................................................................... 7 Pubmed database ....................................................................................................................................................... 7 Redundant sequence database: Nucleotide, Protein, genome, EST… ........................................................................ 8 Performing advances searches using Entrez in other redundant databases ............................................................ 13 Comprehensive database : unigene ......................................................................................................................... 14 comprehensive database Gene ................................................................................................................................ 18 DISCOVERING GENOME PROJECTS IN NCBI VIEW THE GENOME SEQUENCE INITIATIVES (GO TO BIOPROJECT) How many prokaryotic genomes have been sequenced? October 2006: 381 How many are in progress (October 2006: 267)? Note also the nice taxonomic overview of all prokaryotic species that have been sequenced. How many plant species have been fully sequenced What is the 1000 genomes project What is HMP Kathleen Marchal 1 Exercise databases Bioinformatics (updated 2015 january) Can you find the mammoth sequencing project Search for the genomic map of the Chimp link (Pan troglodytes) Microbial genomes Kathleen Marchal 2 Exercise databases Bioinformatics (updated 2015 january) (Browse genomes is at the bottom of the page if you wait long enough) Kathleen Marchal 3 Exercise databases Bioinformatics (updated 2015 january) Plant genomes Kathleen Marchal 4 Exercise databases Bioinformatics (updated 2015 january) Find a full genome (chimp i.e. pan) Kathleen Marchal 5 Exercise databases Bioinformatics (updated 2015 january) GET A LOOK AT THE LARGE INITIATIVES: What is HMP? Why is this an example of a metagenomics project? What is the 1000 genomes project Kathleen Marchal 6 Exercise databases Bioinformatics (updated 2015 january) Why can it be useful See also http://www.1000genomes.org/about The 1000 Genomes Project (human) “The purpose of the project is to support the discovery and understanding of genetic variants that influence human disease. Specifically defined goals are (a) the discovery of single nucleotide variants at frequencies of 1% or higher in diverse populations, (b) even more comprehensive discovery (variants down to frequencies of 0.1 - 0.5%) in functional gene regions, and (c) discovery of structural variants, such as copy number variants, other insertions and deletions, and inversions, including sequence-level understanding of breakpoints. The volume of data generated by 1000genomes project is unprecedented. The data is accessible from two mirrored ftp sites at EBI and NCBI.” USING THE ENTREZ SEARCH ENGINE TO DISCOVER DISTINCT DATABASES AT NCBI PUBMED DATABASE Search for articles on pax6. Kathleen Marchal 7 Exercise databases Bioinformatics (updated 2015 january) REDUNDANT SEQUENCE DATABASE: NUCLEOTIDE, PROTEIN, GENOME, EST… This database contains all the redundant information that is http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=nucleotide used by ENSEMBL and GENE. Search for sequence entries that contain pax6 and human using a simple versus an advanced query 1. First just search for Pax 6, what do you retrieve? 1. Search Pax6 advanced search (organism is human and gene name is pax6) You retrieve many different sequences, these can be mRNAs, genomic DNA,… Kathleen Marchal 8 Exercise databases Bioinformatics (updated 2015 january) Use an advanced search to find pax6 (gene name) in human (organism) and limit to genomic sequences only. There are many entries, most of which only contain part of the sequence (incomplete e.g. only a certain exon and many sequences that come from genomic surveys). Kathleen Marchal 9 Exercise databases Bioinformatics (updated 2015 january) Indicate that you only want the ref seq sequences. 8 entries contains the complete genomic sequence, derived from alternative assemblies. Use the accession number NG_008679.1 to view the genbank file 2. Open the Genbank file of a gene entry and interpret the output (what is the difference between and exon and an mRNA, do you find the Gene ID, this is the link to the comprehensive NCBI database?) Kathleen Marchal 10 Exercise databases Kathleen Marchal Bioinformatics (updated 2015 january) 11 Exercise databases Bioinformatics (updated 2015 january) View the pax 6 graphically Repeat the exercise (Use an advanced search to find pax6 (gene name) in human (organism)) but do now restrict the search to mRNA only. In this case you will find accession numbers that start with NM_: these are REFSEQ sequences or XM_ representing respectively the transcripts or predicted transcripts. Kathleen Marchal 12 Exercise databases Bioinformatics (updated 2015 january) PERFORMING ADVANCES SEARCHES USING ENTREZ IN OTHER REDUNDANT DATABASES 1. This problem practices using the Entrez search program at the national Center for Biotechnology Information (NCBI) to perform a search for the amino acid sequence of the human heat shock factor HSF1. Normally a large number of matches are found in such searches. We will use the Entrez Boolean search features, which restrict the reported matches to a series of required conditions. This feature allows us to narrow the search to the sequence we want. a) Go to the Entrez Web Site and choose Protein b) Enter the terms heat shock factor in the search window and click GO [heat shock protein AND human]. This search is to find any sequence entry in the protein sequence database that include this phrase. c) Now limit the search by clicking the mouse on advanced search, go to add terms, choose organism in the first box, type human in the second, then click AND to limit the search to just human proteins and then click preview. The history will show the results of a search for database entrees with the term heat shock protein AND originating from humans as the organism. How many hits are there now? d) We can limit the hits to matches to RefSeq, which is Genbank’s annotated sequence database, to give a best representative sequence entry for each protein. Click the mouse on Limits, and in the Limited to section of the page, ignore the boxes on the left and choose RefSeq in the right box. Then click GO and history. Now we have all human heat shock factors in RefSeq. The gene of interest is HSF 1. Add this term to the query using. How many hits did you receive? [no limit on gene name] e) The gene of interest is HSF. Click clear in the text entry box at the top of the page, type HSF 1 and click preview. You obtain more hits because you performed a keyword search. It is better to search via the limits option. f) There are other ways of arriving at this final sequence. As another example, pull out all human protein sequences in RefSeq and all HSF 1 sequences in all organisms and then select the human one using another Boolean search feature of Entrez. First clear history, clear the upper text box, and reselect advanced search. Enter human and organism in the text box, click Limits, and limit to RefSEq. Click GO and then History. Now we have a complete list of human sequences in RefSeq. g) Now click Advanced search choose gene name in the left box and HSF1. Combine this search with the previous one using Booleans in the history. The result should be a small number of HSF 1 proteins. Kathleen Marchal 13 Exercise databases Bioinformatics (updated 2015 january) h) Finally note the RefSeq accession number starting with NP and use the mouse click to display the FASTA format. NP identifies the protein as curated protein sequence. The sequence may be copied and pasted into the page of a simple text editor and save as a local computer file. i) While on the page with the target sequence click on LINKs and choose Gene option. Now the gene entry becomes visible. Note that the RefSEq numbers in the GENE database start with NM for annotated mRNA and NT for annotated genome/ chromosome. COMPREHENSIVE DATABASE : UNIGENE Go to Unigene, http://www.ncbi.nlm.nih.gov/unigene/ Go the overview page for human (Unigene statistics). Kathleen Marchal 14 Exercise databases Bioinformatics (updated 2015 january) How many unigene clusters contain only 1 sequence (i.e. unclustered sequences). What will happen if more EST sequences become available. How many clusters contain both an mRNA sequence and an EST. How many only an EST. What will be the most reliable clusters? (HTC = a high throughput cDNA; Sequences in this division may still have 5' and 3' UTRs at their ends, partial coding regions, and introns.) Search for the homo sapiens unigene cluster corresponding to the human pax6. Interpret the output (based on which sequences the cluster was built), Kathleen Marchal 15 Exercise databases Bioinformatics (updated 2015 january) View the expression of the Pax6 gene based on the analysis of EST counts (expression, EST counts). In which tissues do you expect the gene to be expressed? Is this the case? Kathleen Marchal 16 Exercise databases Bioinformatics (updated 2015 january) From the Unigene page Go to the DDD (digital differential display) Compare the difference in expression between two human tissues a) Go to the NCBI Gene. In the first box choose Gene, then Brief in the second and human as search organism in the third, The enter HSF1 as the query and click GO. A small number of entries match the query and one of them should be HSF1. The position column shows the relative numbered position on the long arm (q for the long arm) of chromosome 8. The colored boxes provide sequence of the gene with direct links to RefSeq. Clicking on the green will give the protein sequence. b) Click on the empty check box beside the sequence and then click view to produce a page with a great deal of information about the gene, including gene structure, genome location, RefSEQ protein and nucleotide identifiers AND much useful information about the evidence on which this gene is based. Click on OMIM (online inheritance in man) to see a biological summary of the HSF1 gene functions. Kathleen Marchal 17 Exercise databases Bioinformatics (updated 2015 january) What is the length of the genome, Click on the map to see the genes that are present and then go to the blocks to see the sequences. COMPREHENSIVE DATABASE GENE Check out the Gene database. This is the major curation project at Ncbi. They try to convert the redundant sequence databases into 1 non redundant, comprehensive sequence database in which each locus in the genome is completely described by a representative mRNA sequence(s). Entrez Gene is the American counterpart of ENSEMBL. http://www.ncbi.nih.gov/entrez/query.fcgi?db=gene Search in the gene database for pax6 human (note the difference in result when searching with AND or using the preview/index). Find the accession numbers of the pax6 transcripts, proteins, genome REFSEQ sequence Find the Pax6 gene ID Compare the results with what you found for Pax6 at Ensembl (see later). The Gene database contains for each locus in the genome all associated features (indicated by the corresponding Gene ids). A transcript is indicated by NM (XP if the transcript is still under review), a protein by NP, a genomic contig by NT. All features (mRNA, genomic DNA, EST) associated with the same locus obtain the same Gene ID. The output is less graphical than Ensembl (see below). In Gene non redundant sequence features are also grouped to generate a comprehensive view of the gene. How many transcripts are known?, corresponding to how many different isoforms? Note you find 2 splice variants (now there are more…sequence databases get continuously updated) Find the sequence entries from which Gene was derived. What is the meaning of 2 alternative assemblies? Select the genome view (Can you see the two representative isoforms (red, proteins)?, what is the meaning of the purple squares , variations with a pathogenic phenotype) Kathleen Marchal 18 Exercise databases Bioinformatics (updated 2015 january) Find the GO categories of Pax6 (note there are three ontology classification systems (function, process, component) Find the diseases in which Pax6 is involved Kathleen Marchal 19 Exercise databases Bioinformatics (updated 2015 january) How is this gene found to be related to these diseases? (via GWAS study, how many variants have been detected in this gene? GWAS was performed against which trait?) Kathleen Marchal 20 Exercise databases Kathleen Marchal Bioinformatics (updated 2015 january) 21