* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SNPs - Biology, Genetics and Bioinformatics Unit
Genetic testing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Gene expression programming wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Minimal genome wikipedia , lookup
Gene desert wikipedia , lookup
Transposable element wikipedia , lookup
Genetic code wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genomic library wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Non-coding DNA wikipedia , lookup
Pathogenomics wikipedia , lookup
Frameshift mutation wikipedia , lookup
Copy-number variation wikipedia , lookup
Medical genetics wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Genetic engineering wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Behavioural genetics wikipedia , lookup
Human Genome Project wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic drift wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Microsatellite wikipedia , lookup
Genome editing wikipedia , lookup
Human genome wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Population genetics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Haplogroup G-M201 wikipedia , lookup
Microevolution wikipedia , lookup
SNP genotyping wikipedia , lookup
Public health genomics wikipedia , lookup
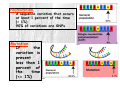
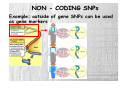
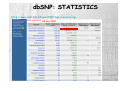
‘The Human Genome Project’ has always been something of a misnomer, implying the existence of a single human genome Of course, every person on the planet with the exception of identical twins has a unique genome, and even though any two genomes are roughly 99.9% identical, identical that still leaves millions of differences among the 3.2 billion base pairs. It is precisely these differences that account for heritable variation among individuals, including susceptibility to disease HUMAN GENETIC VARIATIONS Primarily two types of genetic mutation events create all forms of variations: Single base mutation which substitutes one nucleotide for another -Single Nucleotide Polymorphisms (SNP) Insertion or deletion of one or more nucleotide(s) -Tandem Repeat Polymorphisms -Insertion/Deletion Polymorphisms SINGLE NUCLEOTIDE POLYMORPHISMS Single nucleotide polymorphisms (SNP) are DNA sequence variations that occur when a single nucleotide (A,T,C,or G) in the genome sequence is altered. For example a SNP might change the DNA sequence AAGGCTAA to ATGGCTAA. SNPS are the most common class of polymorphisms. TANDEM REPEAT POLYMORPHISMS Tandem repeats or variable number of tandem repeats (VNTR) are a very common class of polymorphism, consisting of variable length of sequence motifs that are repeated in tandem in a variable copy number. VNTRs are subdivided into two subgroups based on the size of the tandem repeat units. Microsatellites or Short Tandem Repeat (STR) repeat unit: 1-6 (dinucleotide repeat: CACACACACACA) Minisatellites repeat unit: 14-100 example: Spinocerebellar ataxia Type10 (SCA10) (OMIM:+603516) is caused by largest tandem repeat seen in human genome. Normal population has 10-22 mer pentanucleotide ATTCT repeat in intron 9 of SCA10 gene; where as SCA10 patients have 800-4500 repeat units, which causes the disease allele up to 22.5 kb larger than the normal one. INSERTION/DELETION POLYMORPHISMS Insertion/Deletion (INDEL) polymorphisms are quite common and widely distributed throughout the human genome. Sequence repetitiveness in the form of direct or inverted tandem repeat have been shown to predispose DNA to localized rearrangements between homologous repeats. Such rearrangements are thought to be one of the reason which create INDEL polymorphism. example: Association between coronary heart disease and a 287 bp Indel Polymorphism located in intron 16 of the angiotensin converting enzyme (ACE) have been reported (OMIM 106180). This Indel, known as ACE/ID is responsible for 50% of the inter individual variability of plasma ACE concentration. ESTIMATED NUMBERS · SNPs appear at 0.3-1-kb average intervals, considering the size of entire human genome, which is 3X109 bp, the total number scales up to 5-10 million. (Altshuler et al., 2000) · In silico estimation of potentially polymorphic VNTR are over 100,000 across the human genome. · The short insertion/deletions are very difficult to quantify and the number is likely to fall in between SNPs and VNTR VARIATION OR MUTATION ? Terminology for variation at a single nucleotide position is defined by allele frequency. Polymorphism A sequence variation that occurs at least 1 percent of the time (> 1%) 90% of variations are SNPs Mutation If the variation is present less than 1 percent of the time (<= 1%) SINGLE NUCLEOTIDE POLYMORPHISMS (SNPs) SNPs are single base pair positions in genomic DNA at which different sequence alternatives (alleles) exist in normal individuals in some population(s), wherein the least frequent allele has an abundance of 1% or greater. Almost two decades ago the original incarnation of SNPs [as restriction fragment length polymorphisms (RFLPs)] clearly indicated the existence of widespread subtle genome variation ...C C A T T G A C... …G G T A A C T G... ...C C G T T G A C... …G G C A A C T G... LIFE CYCLE OF SNPs AND MUTATIONS TRANSITIONS AND TRANSVERSIONS SNPs include single base substitutions such as: Transitions change of one purine (A,G) for a purine, or a pyrimidine (C,T) for a pyrimidine A G G A C T T C Transversions change of a purine (A,G) for a pyrimidine (C,T), or vice versa A C A T G C G T C A C G T A T G TRANSITIONS AND TRANSVERSIONS The higher level of C>T e G>A SNPs is probably partly related to 5-methylcytosine deamination reactions that are known to occur frequently, particularly at CpG dinucleotides TRANSITIONS AND TRANSVERSIONS In principle, SNPs could be bi-, tri-, or tetraallelic polymorphisms. However, in humans, triallelic and tetra-allelic SNPs are rare almost to the point of non-existence, and so SNPs are sometimes markers simply referred to as bi-allelic CLASSIFICATION OF SNPS SNPs may occur at any position in the above gene structure and based on its location it can be classified as: intronic, exonic or promoter region etc. CLASSIFICATION OF SNPs Non-coding SNPs: 5’ and 3’ UTRs Introns Intergenic Spaces Coding SNPs (subdivided into two groups): Synonymous: when single base substitutions do not cause a change in the resultant amino acid Non-synonymous: when single base substitutions cause a change in the resultant amino acid. NON - CODING SNPs Example: Regulatory SNPs (rSNPs) Two allelic variants of the same gene are transcribed in different amounts as a consequence of an adjacent polymorphism. In this example, allele G, located upstream of the gene, has a higher transcript level than does allele T NON - CODING SNPs Example: outside of gene SNPs can be used as gene markers CODING SNPs Example: Synonymous, mutation does not change amino acid. CODING SNPs Example: Non-synonymous, mutation change amino acid seq. rare mutations that cause medelian diseases with allele frequency below 1%. SNPs DISTRIBUTION 1 SNP per 1 kb sequence, or nucleotide diversity of 10 x 10-4 in the human genome Highest (%) of SNPs Lowest (%) of SNPs The number of SNPs seems to be correlated with the length of the chromosomes The distribution of SNPs among gene structure categories depends on the GC content of the chromosomes SNPs DISTRIBUTION On average, there were 22.59 SNPs per gene and 50.38 SNPs per intergenic regions SNP DATABASES dbSNP http://www.ncbi.nlm.nih.gov/SNP/index.html Human Genome Variation Database (HGVbase) http://hgvbase.cgb.ki.se/ TSC: The SNP Consortium http://snp.cshl.org/ dbSNP URL: http://www.ncbi.nlm.nih.gov/SNP/index.html The Single Nucleotide Polymorphism database (dbSNP) is a public- domain archive for a broad collection of simple genetic polymorphisms. This collection of polymorphisms includes: Single-base nucleotide substitutions (also known as single nucleotide polymorphisms or SNPs) Small-scale multi-base deletions or insertions (also called deletion insertion polymorphisms or DIPs) Microsatellite repeat variations (also called short tandem repeats or STRs). dbSNP: STATISTICS http://www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi 09 Jan 2005 SNP APPLICATIONS • DISEASE MAPPING Direct / Indirect Association Studies • PHARMACOGENOMICS • POPULATION GENETICS DISEASE MAPPING • Linkage Analysis – Within-family associations between marker and putative trait loci • Linkage Disequilibrium (LD) – Across-family associations Linkage and Linkage Disequilibrium (1) • Linkage: the tendency of genes or other DNA sequences at specific loci to be inherited together as a consequence of their physical proximity on a single chromosome. • Linkage disequilibrium (allelic association): particular alleles at two or more neighboring loci show allelic association if they occur together with frequencies significantly different from those predicted from the individual allele frequencies. Linkage is a relation between loci, but association is a relation between alleles. Linkage and Linkage Disequilibrium (2) • Linkage: 0 ≤ θ < 0.5 (θ = recombination fraction) No linkage: θ = 0.5 Perfect linkage: θ = 0 • Linkage disequilibrium: 0 ≤ ρ ≤1 (ρ = probability of allelic association) Linkage equilibrium: ρ = 0 Complete linkage disequilibrium: ρ = 1 Linkage versus Linkage-Disequilibrium Both linkage and linkage-disequilibrium (LD) measures a correlation, or co-segregation, or association, between a genetic marker and the disease affection status 1. Linkage focuses on a locus, Linkage-Disequilibrium focused on an allele 2. Linkage is resulted from recombination events in the last 2-3 generations, Linkage-Disequilibrium is resulted from much earlier, ancestral recombination events 3.Linkage measures co-segregation in a pedigree, Linkage-Disequilibrium measures co-segregation in a population (essentially a huge pedigree) 4. Linkage is usually detected for markers reasonable close to the disease gene (one centiMorgan/one Mb) , Linkage-Disequilibrium is detected for markers even closer (0.01-0.02 centiMorgan/ 10-20 kB). The complexity of common diseases has made them largely refractory to genetic analysis In the face of this complexity, geneticists agree that the family-based approaches that proved so successful for the monogenic diseases are not up to the job Instead, most favor association studies, in which genetic and phenotypic variation is compared in large population samples in order to identify correlations implicating genetic risk factors Association studies compare the allele frequency of a polymorphic marker, or a set of markers, in unrelated patients (cases) and healthy controls to identify markers that differ significantly between the two groups DIRECT ASSOCIATION ANALYSIS Direct association analysis is a direct test of association between a putatively functional variant and disease risk. Example: candidate SNP analysis of coding SNPs (cSNPs) that change amino acids INDIRECT ASSOCIATION ANALYSIS Indirect association is the testing a dense map of SNPs for disease association under the assumption that if a risk polymorphism exists it will either be genotyped directly or be in strong LD with one of the genotyped tagSNPs The advantage of indirect association analysis is that it does not require prior determination of which SNP might be functionally important, but the disadvantage is that a much larger number of SNPs needs to be genotyped 1 2 3 4 5 6 SNP Phenotype Black eye Brown eye Black eye Blue eye Brown eye Brown eye SNP FROM SNP TO HAPLOTYPE GATATTCGTACGGA-T GATGTTCGTACTGAAT GATATTCGTACGGA-T GATATTCGTACGGAAT GATGTTCGTACTGAAT GATGTTCGTACTGAAT Haplotypes AGGTA AGA 2/6 3/6 1/6 DNA Sequence Haplotype: A set of closely linked genetic markers present on one chromosome which tend to be inherited together (not easily separable by recombination). Each person has two haplotypes in a given region, and each haplotype will be passed on as a complete unit C G G A A Set of SNP polymorphisms: a SNP haplotype HAPLOTYPE STUDY Whole-genome genotyping of 10 million SNPs •Technologically daunting •Prohibitively expensive Researchers are trying to downsize the problem of genome-wide genotyping by studying haplotypes. The term genotype can refer to the SNP alleles that a person has at a particular SNP, or for many SNPs across the genome INTERNATIONAL HAP MAP PROJECT The HapMap Home Page URL: http://www.hapmap.org/index.html.en The goal of the International HapMap Project is to develop a haplotype map of the human genome, the HapMap, which will describe the common patterns of human DNA sequence variation. The HapMap will be a tool that will allow researchers to find genes and genetic variations that affect health and disease PHARMACOGENOMICS Pharmacogenomics is a science that examines the inherited variations in genes that dictate drug response and explores the ways these variations can be used to predict whether a patient will have a good response to a drug, a bad response to a drug, or no response at all PHARMACOGENOMICS SNPs in genes encoding drug targets or drug metabolism pathways can determine the therapeutic utility of pharmacologic agents Most drugs show significant interindividual variation in therapeutic efficacy PHARMACOGENOMICS Better, Safer Drugs the First Time: Analyze a patient's genetic profile and prescribe the best available drug therapy from the beginning More Accurate Methods of Determining Appropriate Drug Dosages: Drug dosage can be based on a person's genetics --how well the body processes the medicine and the time it takes to metabolize it. Decrease in the Overall Cost of Health Care: Decrease adverse drug reactions Decrease failed drug trials, Decrease the time for drug approved by government, Decrease the time and the number of medication on patients POPULATION GENETICS Population genetics is the study of the distribution of and change in allele frequencies under the influence of the four evolutionary forces: natural selection, genetic drift, mutation and migration. It also takes account of population subdivision and population structure in space. Genetic polymorphisms can be used to predict the population of origin of an individual Global distribution of genetic diversity. Most genetic variations(shown here as color) is found within individuals of the same population, with a small fraction attributable to differences among populations Population distribution of 37 582 SNPs discovered in 2036 genes. Degree of population sharing is indicated. These differences are expected to have profound consequences for the design of medical association studies and will be of vital importance when trying to identify the genetic contribution to complex phenotypes such as aging Population distribution of distinct haplotypes discovered in 2036 genes. Degree of population sharing is indicated. Though they are small, these differences may be used to partly understand differences in disease risk among populations. The next natural level -differences among individuals- will pave the way for personalized medicine