* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Imam - TU Delft
Vectors in gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
Oncogenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Transposable element wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene desert wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Non-coding DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Metagenomics wikipedia , lookup
Helitron (biology) wikipedia , lookup
Pathogenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
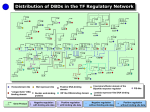
AN INTEGRATED APPROACH TO RECONSTRUCTING GENOME SCALE TRANSCRIPTIONAL REGULATORY NETWORKS SAHEED IMAM ET.AL. A paper pitch by Ivan Valdes, Samar Tareen, Chibuike Ugwuoke, Maryam Suleimani, Gianluca Mazzoni Transcriptional Regulatory Networks • Transcriptionally regulatory networks (TRNs) dynamically alter gene expression in response to stimuli • Many approaches generate TRNs based on the assumption: expression is directly related to cognate transcription factors (TFs). • Drawback: Compromised by indirect effects such as co-expressed but not co-regulated genes • A novel workflow based on: • Integration of comparative genomics data • Global gene expression • Intrinsic properties of TFs The Workflow Phylogenetic Footprinting • Incorporates comparative genomics and phylogenetics • Selection of appropriate organisms • Very closely related organisms: might be uninformative • Very far: might not be conserved Author’s analysis: as few as 6 appropriately selected organisms were sufficient for a robust analysis Identification of Orthologs • Orthologs are genes between species sharing common ancestry • OrthoMCL to detect orthologs • Builds upon the bidirectional best BLAST using Markov Cluster algorithm (MCL) Integration of Gene Expression Data • Matching clusters to expression profiles • Used DISTILLER • Bi-clustering algorithm • Presence/absence of motifs via binary classification • Gives sub-conditions for shared significant coexpression pattern Linking TFs to Clusters • Uses correlation expression profiles to map TFs to targets • Proximity of location • Similarity in DNA binding motif • Phylogenetic correlation QUESTIONS?