* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Life: The Science of Biology, 8e
DNA supercoil wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Human genome wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
RNA interference wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Oncogenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Messenger RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transfer RNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Expanded genetic code wikipedia , lookup
Microevolution wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Frameshift mutation wikipedia , lookup
History of RNA biology wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
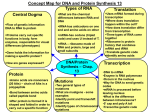
12 From DNA to Protein: Genotype to Phenotype 12.1 What Is the Evidence that Genes Code for Proteins? The gene-enzyme relationship is one-gene, one-polypeptide relationship. Example: In hemoglobin, each polypeptide chain is specified by a separate gene. 12.2 How Does Information Flow from Genes to Proteins? Expression of a gene to form a polypeptide takes 3 main processes: • Transcription—copies information from gene to a sequence of pre-mRNA. • RNA Processing-converts pre-mRNA to mRNA • Translation—converts mRNA sequence to amino acid sequence. 12.2 How Does Information Flow from Genes to Proteins? RNA, ribonucleic acid differs from DNA: • Single strand-so what’s that mean? • The sugar is ribose • Contains uracil (U) instead of thymine (T) 12.2 How Does Information Flow from Genes to Proteins? RNA can pair with a single strand of DNA, except that adenine pairs with uracil instead of thymine. Single-strand RNA can fold into much more unique and differing shapes by internal base pairing. (This flexibility is not seen in DNA) Figure 12.2 The Central Dogma The central dogma of molecular biology for eukaryotes: information flows in one direction when genes are expressed (Francis Crick). 12.2 How Does Information Flow from Genes to Proteins? ONE Exception to the central dogma: Viruses: acellular particles that reproduce inside cells; many have RNA instead of DNA so reverse the process. Synthesis of DNA from RNA is called reverse transcription. Viruses that do this are called retroviruses 12.2 How Does Information Flow from Genes to Proteins? Messenger RNA (mRNA) forms as a complementary copy of DNA and carries information to the cytoplasm. (WHY use a copy of DNA?) This process is called transcription and occurs in the nucleus. RNA polymerase is the enzyme that runs the same direction as it’s “cousin”. Will we have a leading or lagging strand now? Why or why not? Figure 12.3 From Gene to Protein 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? Transcription occurs in three phases: • Initiation • Elongation • Termination 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? Initiation requires a promoter—a special sequence of DNA. RNA polymerase binds to the promoter. Promoter tells RNA polymerase where to start, which direction to go in, and which strand of DNA to transcribe. In eukaryotes it is the “TATA” region called the initiation site. Figure 12.5 DNA Is Transcribed to Form RNA (A) 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? Elongation: RNA polymerase copies base pairs of DNA into pre-mRNA. RNA polymerase also runs in a 5-3 direction. (So what DNA template will we use? Why? What about the other one?) Figure 12.5 DNA Is Transcribed to Form RNA (B) 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? Termination: specified by a specific DNA base sequence. Mechanisms of termination are complex and varied. Figure 12.5 DNA Is Transcribed to Form RNA (C) Eukaryotes—first product is a premRNA that is longer than the final mRNA and must undergo processing. The Pre mRNA must be readied for travel so 5’ caps and poly A tails (3’) are added to the strand. Non coding regions called introns are also removed leaving only exons. Once RNA processing is complete, we have mRNA • Please get a book and turn to page 262 Before we begin, we need to understand that RNA is extremely flexible! • There are 4 types of RNA, each encoded by its own type of gene: • mRNA - Messenger RNA: Encodes amino acid sequence of a polypeptide. • tRNA - Transfer RNA: Brings amino acids to ribosomes during translation. • rRNA - Ribosomal RNA: With ribosomal proteins, makes up the ribosomes, the organelles that translate the mRNA. • snRNA - Small nuclear RNA: With proteins, forms complexes that are used in RNA processing in eukaryotes. (Not found in prokaryotes.) (This is what splices out introns!) 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? The genetic code: specifies which amino acids will be used to build a protein Codon: a sequence of three bases. Each codon specifies a particular amino acid. Start codon: AUG—initiation signal for translation Stop codons: stops translation and polypeptide is released-UAA, UGA or UAG Figure 12.6 The Genetic Code 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? How do we keep from having too many mutations? For most amino acids, there is more than one codon; the genetic code is redundant. How does that protect the integrity of proteins? 12.3 How Is the Information Content in DNA Transcribed to Produce RNA? The genetic code is nearly universal: the codons that specify amino acids are the same in all organisms. That means we get uniqueness because of the sequence of amino acids Even in all that diversity, all life uses the same start and stop codons! 12.4 How Is RNA Translated into Proteins? Let’s look at each type of RNA now…. Functions of tRNA: • Carries an inactive amino acid • Carries an active amino acid • Interacts with ribosomes by providing the anticodon Figure 12.8 Transfer RNA 12.4 How Is RNA Translated into Proteins? The conformation (three-dimensional shape) of tRNA results from base pairing (H bonds) within the molecule. Anticodon: site of base pairing with mRNA. Unique for each species of tRNA. Formula for building a protein is Codon + anticodon + inactive aa= specific aa in polypeptide chain 12.4 How Is RNA Translated into Proteins? Example: DNA codon for alanine: GCC Complementary mRNA: CGG Anticodon on the tRNA: GCC Active amino acid would be: alanine 12.4 How Is RNA Translated into Proteins? Wobble: specificity for the base on tRNA so one tRNA can decode up to 3 different codons. Example: codons for alanine—GCA, GCC, and GCU—are recognized by the same tRNA. Allows cells to produce fewer tRNA. 12.4 How Is RNA Translated into Proteins? Ribosome: the workbench—holds mRNA and tRNA in the correct positions to allow assembly of polypeptide chain. Ribosomes are not specific, they can make any type of protein. 12.4 How Is RNA Translated into Proteins? rRNA: AKA the Ribosomes have two subunits, large and small. The subunits are made of rRNA or ribosomal RNA. Figure 12.10 Ribosome Structure 12.4 How Is RNA Translated into Proteins? Large subunit has three tRNA binding sites: • A site binds with anticodon of charged tRNA. Activation • P site is where tRNA adds its amino acid to the growing chain. Polypeptide chain is held and built • E site is where tRNA sits before being released. Exit 12.4 How Is RNA Translated into Proteins? Translation also occurs in three steps: • *Initiation-start codon (AUG) first amino acid is always methionine • Elongation of the polypeptide chain • Termination- stop codon enters the A site. Methionine (AUG) hits the P site of the small ribosomal sub-unit that action initiates the process. One of the first things that happens is the large ribosomal sub-unit joins with the small unit and makes an rRNA Figure 12.11 The Initiation of Translation (Part 1) Figure 12.11 The Initiation of Translation (Part 2) Figure 12.12 The Elongation of Translation (Part 1) Figure 12.12 The Elongation of Translation (Part 2) Figure 12.13 The Termination of Translation (Part 1) Figure 12.13 The Termination of Translation (Part 2) Figure 12.13 The Termination of Translation (Part 3) Table 12.1 Figure 12.14 A Polysome (Part 1) Figure 12.14 A Polysome (Part 2) • http://highered.mheducation.com/sites/ 0072507470/student_view0/chapter3/a nimation__how_translation_works.html • http://www.stolaf.edu/people/giannini/fla shanimat/molgenetics/translation.swf • http://www.phschool.com/science/biolog y_place/biocoach/transcription/difgns.ht ml Figure 12.15 Destinations for Newly Translated Polypeptides in a Eukaryotic Cell 12.6 What Are Mutations? Somatic mutations occur in somatic (body) cells. Mutation is passed to daughter cells, but not to sexually produced offspring. Germ line mutations occur in cells that produce gametes. Can be passed to next generation. This is the key to evolution and are available to occur in transcription. 12.6 What Are Mutations? All mutations are alterations of the nucleotide sequence. 2 levels of mutation…. Point mutations: change in a single base pair—loss, gain, or substitution of a base. Chromosomal mutations: change in segments of DNA—loss, duplication, or rearrangement. 12.6 What Are Mutations? Point mutations can result from replication and proofreading errors, or from environmental mutagens. Silent mutations have no effect on the protein because of the redundancy of the genetic code. Silent mutations result in genetic diversity not expressed as phenotype differences. 12.6 What Are Mutations? 12.6 What Are Mutations? KEY! These CAN be beneficial! Missense mutations: base substitution results in amino acid substitution. 12.6 What Are Mutations? Sickle allele for human β-globin is a missense mutation. Sickle allele differs from normal by only one base—the polypeptide differs by only one amino acid. Individuals that are homozygous have sickle-cell disease. Figure 12.18 Sickled and Normal Red Blood Cells 12.6 What Are Mutations? Nonsense mutations: base substitution results in a stop codon. 12.6 What Are Mutations? Frame-shift mutations: single bases inserted or deleted—usually leads to nonfunctional proteins. 12.6 What Are Mutations? Chromosomal mutations: Deletions—severe consequences unless it affects unnecessary genes or is masked by normal alleles. Duplications—if homologous chromosomes break in different places and recombine with the wrong partners. Figure 12.19 Chromosomal Mutations (A, B) 12.6 What Are Mutations? Chromosomal mutations: Inversions—breaking and rejoining, but segment is “flipped.” Translocations—segment of DNA breaks off and is inserted into another chromosome. Can cause duplications and deletions. Meiosis can be prevented if chromosome pairing is impossible. Figure 12.19 Chromosomal Mutations (C, D) 12.6 What Are Mutations? • Replication errors—some escape detection and repair. • Nondisjunction in meiosis. 12.6 What Are Mutations? Mutation provides the raw material for evolution in the form of genetic diversity. Mutations can harm the organism, or be neutral. Occasionally, a mutation can improve an organism’s adaptation to its environment, or become favorable as conditions change. Eukaryotic gene regulationTATA REGION=3'-TATAAT-5’ RNA PROCESSING 12.6 What Are Mutations? Induced mutation—due to an outside agent, a mutagen. Chemicals can alter bases Prokaryotic gene regulation much simpler! Operons are repeating regions that make up the prokaryote’s genome They include 3 main regions; promoter, operator and structural genes The 2 main operon examples are the inducible (lac for lactose) or repressible (trp for tryptophan) models What do you think regulatory genes code for? Lac operon structural genes, produce enzymes that can digest lactose. So, when would you want to transcribe and translate those enzymes? In the lac (lactose) model, we call this method of gene regulation “inducible” as the substance (lactose) causes the genes to be on or off. (Lactose induces the operon to be on) Let’s try it! Open notebook Since lactose is a factor in the operating of this enzyme in that it Changes the repressor protein’s shape so it can not fit into the operator region, we call it “allolactose”. But what about if the mRNA we make creates proteins that make A substance we need? Tryptophan is an essential substance for bacterial metabolism. Therefore when would you want the operon “ON” and the creation Of tryptophan to occur? Let’s try it… Open notebook What happens to levels of trp as we go on and on? Yes! As more and more trp is made, enough already! So, Some of the trp made binds to the inactive repressor to change it’s shape and allow it to bind to the promoter! The product (trp) is a co-repressor when levels get high enough! Complete this table: Lac on when: Lac off when: So what determines? Trp on when: Trp off when: So what determines?