* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Subcellular organelles in Eukaryotic cells
Survey
Document related concepts
Cytoplasmic streaming wikipedia , lookup
Cell growth wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell culture wikipedia , lookup
Lipid bilayer wikipedia , lookup
Model lipid bilayer wikipedia , lookup
Cell nucleus wikipedia , lookup
Extracellular matrix wikipedia , lookup
Tissue engineering wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cell encapsulation wikipedia , lookup
Type three secretion system wikipedia , lookup
Cytokinesis wikipedia , lookup
Lipopolysaccharide wikipedia , lookup
Signal transduction wikipedia , lookup
Cell membrane wikipedia , lookup
Transcript
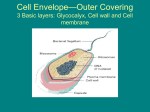
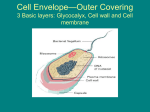
KAIST Subcellular organelles in Eukaryotic cells Biomolecular Engineering Mitochondria Mitochondria are shaped like sausages, and contains two membranes. Mitochondria also contain their own DNA (transferred from mother to their offspring) often called the powerhouse of the cell because they make energy for the cell. They produce energy by turning ADP into ATP in inner membrane. Cells that need more energy, such as muscle cells, contain more mitochondria than cells that need less energy. Ch1 The Development of Molecular Biotechnology KAIST Subcellular organelles in Eukaryotic cells Biomolecular Engineering The Endoplasmic p Reticulum (ER) An interconnected system of flattened, double-layered membranes. The ER is connected to the nuclear membrane. Some parts of the ER contain many ribosomes on their surfaces, giving them a rough appearance (called rough ER) ER), Parts of the ER without ribosomes are called smooth ER ER. Ribosome are responsible for proteins synthesis through translation process, so in eukaryotic cells, translation occurs on the surface of ER not within nuclues. Synthesized proteins are delivered to Golgi body in which they move to other parts of cells. So ER is called mail-man. Ribosomes Ch1 The Development of Molecular Biotechnology KAIST Subcellular organelles in Eukaryotic cells Biomolecular Engineering The Golgi apparatus, apparatus also called Golgi body or Golgi complex, complex It looks like a set of flattened sacs. The Golgi apparatus packages and transports materials, which are then carried in vesicles through the cytoplasm to other parts of the cell, or are excreted from the cell (Secretion process). Its nickname is Post office. Many post-translational modification for proteins occurs during transports through ER and Golgi apparatus Ch1 The Development of Molecular Biotechnology KAIST Subcellular organelles in Eukaryotic cells Biomolecular Engineering Lysosomes y a special type of vesicle that contain digestive enzymes. Round structures surrounded by membranes. Lysosomes recycle materials by breaking down worn-out parts of a cell into smaller units (sweeper in a cell) cell). They deliver these small materials to the cytoplasm for use in constructing new proteins (recycling). If the membrane of a lysosome breaks, the enzymes released may also destroy the cell itself (autolysis), giving lysosomes the name "suicide bag". Ch1 The Development of Molecular Biotechnology KAIST Subcellular organelles in Eukaryotic cells Biomolecular Engineering Ribosomes are produced in the nucleolus (inside Nucleus). Nucleus) Ribosomes are found on the outside of the ER, as well as floating free in the cytoplasm. Ribosomes contain a small and large subunit, and consist of proteins and rRNA. Their function is to synthesize proteins by assembling amino acids according to instructions from mRNA (Translation). Translation by Ribosome (in Chapter 2) Ch1 The Development of Molecular Biotechnology KAIST Plant cells Biomolecular Engineering Vacuoles are membrane-bound sacs within the cytoplasm of a cell that function in several different ways. In mature plant l t cells, ll vacuoles l tend t d to t be b very large l andd are extremely t l important i t t in i providing idi structural t t l support, t as well ll as serving i functions such as storage, waste disposal, protection, and growth. Many plant cells have a large, single central vacuole that typically takes up most of the room in the cell (80 percent or more). Vacuoles in animal cells, however, tend to be much smaller, and are more commonly used to temporarily store materials or to transport substances. The central vacuole ac ole in i plant l t cells ll is i enclosed l d by b a membrane b termed t d the th tonoplast, tonoplast an important i t t andd highly hi hl integrated i t t d component of the plant internal membrane network (endomembrane) system. Ch1 The Development of Molecular Biotechnology KAIST Chloroplast (엽록체) Chloroplasts Contains chlorophyll (inside thylakoid membrane), hence provide the green color. color Double membranes (inner and outer). Has its own DNA which codes for redox proteins involved in electron transport in photosynthesis Thylakoid, Stroma, Granum, lumen are included Photosynthesis takes place on the thylakoid membrane. Ch1 The Development of Molecular Biotechnology Biomolecular Engineering KAIST Classification of Bacteria According to various criteria Shape, Shape Gram staining, staining cellular nutrition & energy metabolism O2 requirement Ch1 The Development of Molecular Biotechnology Biomolecular Engineering KAIST Gram staining Biomolecular Engineering Gram staining : Bacterial cell staining method developed by the Danish scientist Hans Christian Gram (1853 – 1938) in 1884 Principle : Different permeability of the cell wall to “purple colored iodine dye complex” Brief procedure 1) Fixation of the cell on the slide glass 2) Staining with crystal violet 3) Staining with iodine (potassium iodide) Crystal violet and iodine treatment produces "purple colored iodine-dye complexes" inside bacteria (cytoplasm). 4) Treatment with a decolonizing agent such as 95% ethanol or a mixture of acetone and alcohol Under this treatment, the cell wall of Gram (-) cells are permeable to "purple colored iodine-dye complexes“, so in this step, Gram (-) cells are decolorized. 5) To visualize decolorized Gram (-) bacteria, a red counterstain such as safranin is used after decolorization treatment treatment. Ch1 The Development of Molecular Biotechnology Gram staining KAIST Gram (+) Purple color (Bacillus anthracis) Ex) Bacillus sp., Staphylococcus sp., Listeria sp., Streptococcus sp., Clostridium sp., Enterococcus sp. Biomolecular Engineering Gram (-) Red color (Escherichia coli) Ex) Escherichia coli, Salmonella, Shigella, and other Enterobacteriaceae, Pseudomonas, Helicobacter, Bdellovibrio, Legionella Cyanobacteria, green sulfur and green nonsulfur bacteria. Ch1 The Development of Molecular Biotechnology KAIST Gram staining Biomolecular Engineering Much thicker (multilayered) peptideglycan structure Only one cytoplasmic membrane (lipid bilayer) No periplasmic space Lipoteicoic acid (LTA) are found only y in Gram ((+)) Thin peptidoglycan structure Two membranes (inner and outer – same lipid bilayer) Periplasmic space between two membranes. e b a es. Lipopolyscharide (LPS) are found only in Gram (-) Ch1 The Development of Molecular Biotechnology KAIST Peptidoglycan Biomolecular Engineering Peptidoglycan ept dog yca : Polymer o y e by ccrosslinking oss g betwee between N-Acetylglucosamin N cety g ucosa and a d N-Acetylmuramic N cety u a c acid ac d The structure of the polysaccharide backbone of peptidoglycan is conserved in all bacteria, with only minor variations. N-acetylglucosamin N-acetylmuramic acid Lysozyme : peptidoglycan-lytic enzyme (from egg white) Lysozyme y y Loss of Rigidity & Cell lysis Ch1 The Development of Molecular Biotechnology KAIST Lipid bilayer Biomolecular Engineering Gram (-) bacteria Phospholipid p p Major components of membrane lipid Amphiphatic lipid OM Peptidoglycan Periplasm IM Cytoplasm Ch1 The Development of Molecular Biotechnology Lipid bilayer KAIST Biomolecular Engineering Phospholipid p p Gram (+) bacteria Major components of membrane lipid Amphiphatic lipid Peptidoglycan IM Cytoplasm Ch1 The Development of Molecular Biotechnology KAIST Summary of Gram (-) bacteria Biomolecular Engineering Two membranes b (inner (i andd outer membranes) b ) consisting i i off lipid li id bilayers bil Thin peptidoglycan layer between two membranes Outer membrane containing lipopolysaccharide (LPS, which consists of lipid A, core polysaccharide, and O antigen) outside the peptidoglycan layer Periplasmic space between the plasma membrane and the outer membrane Example species Escherichia coli, Salmonella sp., Pseudomonas sp., Shigella sp., Helicobacter sp., Bdellovibrio sp., Legionella sp., Cyanobacteria etc LPS Outer membrane Extracellular medium Peptidoglycan layer Periplasm Chromosome Cytoplasm Inner membrane Flagella Pili Ch1 The Development of Molecular Biotechnology KAIST Escherichia coli Biomolecular Engineering Escherichia coli Nonpathogenic, short, motile, rod-shaped organism Cell length: about ~3 μm, diameter: about ~1 μm One chromosome – full sequencing was completes in 1997 (4.3 Mb). Naturally found in the intestines of humans Optimal temperature for cell growth : 37oC, One generation time (doubling time) is about 22 min (The shortest time in all bacteria). Aerobically or anaerobically: grown aerobically for the optimal production of recombinant protein (Facultative anaerobe) b ) •The most important/widely used host strain in commercial production of recombinant proteins/metabolites The fastest growth High cell density cultivation in large scale culture Easy manipulation of cells and many genetic engineering tools l Deep understanding physiology/metabolism Ch1 The Development of Molecular Biotechnology Summary of Gram (+) bacteria KAIST Biomolecular Engineering Gram-positive p cell Have much thicker peptidoglycan layers in cell walls than Gram-negative and so are able to retain a crystal violet dye during the Gram stain process. Do not have an outer membrane, but rather they have a very thick, rigid cell wall (15-80 nm) with multiple layers of peptidoglycan Have only a cytoplasmic membrane (No periplasm) Often much better suited to secretion of proteins • Example species Bacillus sp., Staphylococcus sp., Listeria sp., Streptococcus sp., Clostridium sp., Enterococcus sp. Peptidoglycan layer Extracellular medium Cytoplasm Cytoplasmic membrane Chromosome Flagella Pilus Ch1 The Development of Molecular Biotechnology KAIST Eukaryotic cells - Yeast Biomolecular Engineering Saccharomyces y cerevisiae (In chapter p 7) Nonpathogenic, single-celled (~ 5 um diameter) yeast Sugar ethanol & CO2: used for the production of alcoholic beverage & bread Complete set (16 pairs) of chromosomes entirely sequenced (1996) first eukaryotic organism Useful host for the production of recombinant proteins from eukaryotic organisms Eukaryotic version of E. coli - widely used for the production of recombinant proteins Other host yeasts for heterologous proteins : Pichia pastoris & Hansenula polymorpha Post-translational modification Enzymatic addition of compounds to proteins after their synthesis (Translation) in eukaryotic organisms Glycosylation (sugar), phosphorylation (Phosphate), acetylation (acetic acid), sulfation (Sulfur) etc Essential for the proper functioning of the protein in some proteins EPO, EPO hTPO (human thrombopoietin) E. coli and other prokaryotes No ability to perform the protein modifications Ch1 The Development of Molecular Biotechnology Eukaryotic cells - Yeast KAIST Glycosylation Human Biomolecular Engineering Bacteria N-Glycosylated protein in higher organisms In bacterial host (Escherichia coli) In human or Yeast Ch1 The Development of Molecular Biotechnology