* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gill: Transcription Regulation I
Deoxyribozyme wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene therapy wikipedia , lookup
RNA silencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Pathogenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Genetic engineering wikipedia , lookup
Transposable element wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genomic library wikipedia , lookup
Point mutation wikipedia , lookup
Gene desert wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Human genome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Non-coding RNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Non-coding DNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
CS273A
Lecture 5: Transcription Regulation I
MW 12:50-2:05pm in Beckman B302
Profs: Serafim Batzoglou & Gill Bejerano
TAs: Harendra Guturu & Panos Achlioptas
http://cs273a.stanford.edu [BejeranoFall13/14]
1
Announcements
• HW1 is out. Due by 11.00 AM Friday, October 18.
–Check it out.
http://cs273a.stanford.edu [BejeranoFall13/14]
2
ATATTGAATTTTCAAAAATTCTTACTTTTTTTTTGGATGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA
TATCCATATCTAATCTTACTTATATGTTGTGGAAATGTAAAGAGCCCCATTATCTTAGCCTAAAAAAACCTTCTCTTTGGAACTTTC
TAATACGCTTAACTGCTCATTGCTATATTGAAGTACGGATTAGAAGCCGCCGAGCGGGCGACAGCCCTCCGACGGAAGACTCTCCTC
TGCGTCCTCGTCTTCACCGGTCGCGTTCCTGAAACGCAGATGTGCCTCGCGCCGCACTGCTCCGAACAATAAAGATTCTACAATACT
CTTTTATGGTTATGAAGAGGAAAAATTGGCAGTAACCTGGCCCCACAAACCTTCAAATTAACGAATCAAATTAACAACCATAGGATG
AATGCGATTAGTTTTTTAGCCTTATTTCTGGGGTAATTAATCAGCGAAGCGATGATTTTTGATCTATTAACAGATATATAAATGGAA
GCTGCATAACCACTTTAACTAATACTTTCAACATTTTCAGTTTGTATTACTTCTTATTCAAATGTCATAAAAGTATCAACAAAAAAT
TTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAATGACTAAATCTCATTCAGAAGAAGTGATTGTACCTGAGTTCAA
CTAGCGCAAAGGAATTACCAAGACCATTGGCCGAAAAGTGCCCGAGCATAATTAAGAAATTTATAAGCGCTTATGATGCTAAACCGG
TTTGTTGCTAGATCGCCTGGTAGAGTCAATCTAATTGGTGAACATATTGATTATTGTGACTTCTCGGTTTTACCTTTAGCTATTGAT
TGATATGCTTTGCGCCGTCAAAGTTTTGAACGATGAGATTTCAAGTCTTAAAGCTATATCAGAGGGCTAAGCATGTGTATTCTGAAT
TTAAGAGTCTTGAAGGCTGTGAAATTAATGACTACAGCGAGCTTTACTGCCGACGAAGACTTTTTCAAGCAATTTGGTGCCTTGATG
CGAGTCTCAAGCTTCTTGCGATAAACTTTACGAATGTTCTTGTCCAGAGATTGACAAAATTTGTTCCATTGCTTTGTCAAATGGATC
ATGGTTCCCGTTTGACCGGAGCTGGCTGGGGTGGTTGTACTGTTCACTTGGTTCCAGGGGGCCCAAATGGCAACATAGAAAAGGTAA
GAAGCCCTTGCCAATGAGTTCTACAAGGTCAAGTACCCTAAGATCACTGATGCTGAGCTAGAAAATGCTATCATCGTCTCTAAACCA
ATTGGGCAGCTGTCTATATGAATTAGTCAAGTATACTTCTTTTTTTTACTTTGTTCAGAACAACTTCTCATTTTTTTCTACTCATAA
TTAGCATCACAAAATACGCAATAATAACGAGTAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGA
ATGTTTTCAATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTT
ATACCTATTCTTGACATGATATGACTACCATTTTGTTATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTTGCGAAGTT
TGGCAAGTTGCCAACTGACGAGATGCAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGT
TCAATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTTGGATAC
ATTCTTGACATGATATGACTACCATTTTGTTATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTCATTTGCGAAGTTCT
GCAAGTTGCCAACTGACGAGATGCAGTTTCCTACGCATAATAAGAATAGGAGGGAATATCAAGCCAGACAATCTATCATTACATTTA
CGGCTCTTCAAAAAGATTGAACTCTCGCCAACTTATGGAATCTTCCAATGAGACCTTTGCGCCAAATAATGTGGATTTGGAAAAAGA
ATAAGTCATCTCAGAGTAATATAACTACCGAAGTTTATGAGGCATCGAGCTTTGAAGAAAAAGTAAGCTCAGAAAAACCTCAATACA
TCATTCTGGAAGAAAATCTATTATGAATATGTGGTCGTTGACAAATCAATCTTGGGTGTTTCTATTCTGGATTCATTTATGTACAAC
GGACTTGAAGCCCGTCGAAAAAGAAAGGCGGGTTTGGTCCTGGTACAATTATTGTTACTTCTGGCTTGCTGAATGTTTCAATATCAA
CTTGGCAAATTGCAGCTACAGGTCTACAACTGGGTCTAAATTGGTGGCAGTGTTGGATAACAATTTGGATTGGGTACGGTTTCGTTG
GCTTTTGTTGTTTTGGCCTCTAGAGTTGGATCTGCTTATCATTTGTCATTCCCTATATCATCTAGAGCATCATTCGGTATTTTCTTC
TTTATGGCCCGTTATTAACAGAGTCGTCATGGCCATCGTTTGGTATAGTGTCCAAGCTTATATTGCGGCAACTCCCGTATCATTAAT
TGAAATCTATCTTTGGAAAAGATTTACAATGATTGTACGTGGGGCAGTTGACGTCTTATCATATGTCAAAGTCATTTGCGAAGTTCT
GCAAGTTGCCAACTGACGAGATGCAGTAACACTTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTT
AATGTAAGAGATTTCGATTATCCACAAACTTTAAAACACAGGGACAAAATTCTTGATATGCTTTCAACCGCTGCGTTTTGGATACCT
TCTTGACATGATATGACTACCATTTTGTTATTGTTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTT
AATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGA
TTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTA
CTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTT
TACATGCTTCAACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTT
ACTACTTAATAAATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAA
http://cs273a.stanford.edu [BejeranoFall13/14]
3
AATGATTGTATGATAATGTTTTCAATGTAAGAGATTTCGATTATCCTTATAGTTCATACATGCTTCAACTACTTAATAAATGATTGT
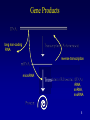
Gene Products
long non-coding
RNA
reverse transcription
microRNA
rRNA,
snRNA,
snoRNA
4
Gene Regulatory Switches
• Gene = genomic substring that encodes
HOW to make a protein (or ncRNA).
• Genomic switch = genomic substring that encodes
WHEN, WHERE & HOW MUCH of a protein to make.
[0,1,1,1]
B
H
Gene
Gene
N
B
N
H
Gene
Gene
http://cs273a.stanford.edu [BejeranoFall13/14]
[1,0,0,1]
[1,1,0,0]
5
If you only measure gene expression
It’s like only seeing the
values change in RAM
as a program is running.
http://cs273a.stanford.edu [BejeranoFall13/14]
6
Cis (=close) regulatory elements
Type
CIS REGULATION
# in genome
% of genome
genes
25,000
2%
ncRNA
15,000
1%
1,000,000
>10%
cis elements
•
•
•
•
•
Encode causality
Disease susceptibility
Driver sequences
Alter cell state
Key for evolution
promoters, enhancers, silencers, insulators
http://cs273a.stanford.edu [BejeranoFall13/14]
7
Transcription Activation
http://cs273a.stanford.edu [BejeranoFall13/14]
8
RNA Polymerase
• Transcription = Copying a segment of DNA into (non/coding) RNA
• Gene transcription starts at the (aptly named) TSS, or
gene transcription start site
• Transcription is done be RNA polymerase, a complex of 10-12
subunit proteins.
• There are three types of RNA polymerases in human:
– RNA pol I synthesizes ribosomal RNAs
– RNA pol II synthesizes pre-mRNAs and most microRNAs
– RNA pol III synthesizes tRNAs, rRNA and other ssRNAs
TSS
RNA Polymerase
http://cs273a.stanford.edu [BejeranoFall13/14]
9
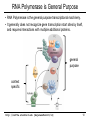
RNA Polymerase is General Purpose
• RNA Polymerase is the general purpose transcriptional machinery.
• It generally does not recognize gene transcription start sites by itself,
and requires interactions with multiple additional proteins.
general
purpose
context
specific
http://cs273a.stanford.edu [BejeranoFall13/14]
10
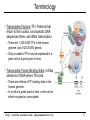
Terminology
• Transcription Factors (TF): Proteins that
return to the nucleus, bind specific DNA
sequences there, and affect transcription.
– There are 1,200-2,000 TFs in the human
genome (out of 20-25,000 genes)
– Only a subset of TFs may be expressed in a
given cell at a given point in time.
• Transcription Factor Binding Sites: 4-20bp
stretches of DNA where TFs bind.
– There are millions of TF binding sites in the
human genome.
– In a cell at a given point in time, a site can be
either occupied or unoccupied.
http://cs273a.stanford.edu [BejeranoFall13/14]
11
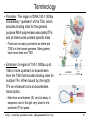
Terminology
• Promoter: The region of DNA 100-1,000bp
immediately “upstream” of the TSS, which
encodes binding sites for the general
purpose RNA polymerase associated TFs,
and at times some context specific sites.
– There are as many promoters as there are
TSS’s in the human genome. Many genes
have more than one TSS.
• Enhancer: A region of 100-1,000bp up to
1Mb or more upstream or downstream
from the TSS that includes binding sites for
multiple TFs. When bound by (the right)
TFs an enhancer turns on/accelerates
transcription.
– Note how an enhancer (E) very far away in
sequence can in fact get very close to the
promoter (P) in space.
http://cs273a.stanford.edu [BejeranoFall13/14]
promoter
TSS
gene
12
TFBS Position Weight Matrix (PWM)
Note the strong independence assumption between positions.
Holds for most transcription binding profiles in the human genome.
http://cs273a.stanford.edu [BejeranoFall13/14]
13
Promoters
http://cs273a.stanford.edu [BejeranoFall13/14]
14
Enhancers
http://cs273a.stanford.edu [BejeranoFall13/14]
15
Terminology
• Gene regulatory domain: the full repertoire
of enhancers that affect the expression of a
(protein coding or non-coding) gene, at
some cells under some condition.
promoter
– Gene regulatory domains do not have to be
contiguous in genome sequence.
– Neither are they disjoint: One or more
enhancers may well affect the expression of
multiple genes (at the same or different times).
TSS
http://cs273a.stanford.edu [BejeranoFall13/14]
enhancers for different contexts
16
Imagine a giant state machine
Transcription factors bind DNA, turn on or off different promoters and
enhancers, which in-turn turn on or off different genes, some of which
may themselves be transcription factors, which again changes the
presence of TFs in the cell, the state of active promoters/enhancers etc.
Proteins
DNA
transcription factor
binding site
Gene
DNA
http://cs273a.stanford.edu [BejeranoFall13/14]
17
One nice hypothetical example
requires active enhancers to function
functions independently of enhancers
http://cs273a.stanford.edu [BejeranoFall13/14]
18
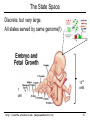
The State Space
Discrete, but very large.
All states served by same genome(!)
1
cell
http://cs273a.stanford.edu [BejeranoFall13/14]
1012
cells
19
Transcription Activation:
Some measurements and observations
http://cs273a.stanford.edu [BejeranoFall13/14]
20
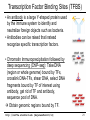
Transcription Factor Binding Sites (TFBS)
• An antibody is a large Y-shaped protein used
by the immune system to identify and
neutralize foreign objects such as bacteria.
• Antibodies can be raised that instead
recognize specific transcription factors.
• Chromatin Immunoprecipitation followed by
deep sequencing (ChIP-seq): Take DNA
(region or whole genome) bound by TFs,
crosslink DNA-TFs, shear DNA, select DNA
fragments bound by TF of interest using
antibody, get rid of TF and antibody,
sequence pool of DNA.
Obtain genomic regions bound by TF.
http://cs273a.stanford.edu [BejeranoFall13/14]
21
ChIP-seq Position Weight Matrix
Computational challenge:
The sequenced DNA
fragments are 200-500bp.
In each is one or more
instance of the 6-20bp motif.
Find it…
http://cs273a.stanford.edu [BejeranoFall13/14]
22
Transcription Factors have Large “fan outs”
We could have had one TF regulate two TFS, each of
which regulates two other TFs, etc. and each of those
contributing to the regulation of a modest number of target
genes (that do the real work).
Instead TFs reproducibly bind to thousands of genomic
locations almost anywhere we’ve looked.
Gene regulation forms a dense network.
http://cs273a.stanford.edu [BejeranoFall13/14]
23
Transfections
As far as we’ve seen, enhancers work “the same” irrespective
of distance (or orientation) to TSS, or identity of target gene.
enhancer
reporter gene
minimal
promoter
in cellular
context
of choice
• Which enhancers work in what contexts?
• What if you mutate enhancer bases
(disrupt or introduce binding sites)
and run the experiment again?
• What if you co-transfect a TF you think
binds to this enhancer?
• What if you instead add siRNA for that TF?
http://cs273a.stanford.edu [BejeranoFall13/14]
24
Transcription factors bind synergistically,
often with preferred spacing
Transcription factor complexes
prefer specific spacings!
Sox:1 bp:Pax
Sox2
Pax6
Sox2 Pax6
0
5
10
60
80
100 120
Fold activation
140
160
180
0
5
10
60
80
100 120
Fold activation
140
160
180
{+2}
Sox:3 bp:Pax
Sox2
Pax6
Sox2
Pax6
Adapted from Kamach et al., Genes Dev, 2001
http://cs273a.stanford.edu [BejeranoFall13/14]
25
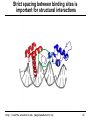
Strict spacing between binding sites is
important for structural interactions
http://cs273a.stanford.edu [BejeranoFall13/14]
26
Complexes may leave genomic footprints
• If a complex prefers
TF : spacer : TF
• This pattern may be
abundant in the
genome
TAAACAGGAAGT
AAAACAGGAATA
ATAACAGGATGC
TTAACAGGAAAG
TAAACAGGATAG
AAAACAGGAAAA
Can we read complexes from individual predictions?
http://cs273a.stanford.edu [BejeranoFall13/14]
27
Cooperative binding of complexes can be
detected as the co-occurrence of individuals
Each dot = different spacer
http://cs273a.stanford.edu [BejeranoFall13/14]
28
Co-occurrences can be filtered for only
structurally feasible patterns
=
=
Fox { spacer } Ets
Remove physically
incompatible configurations
compatible
http://cs273a.stanford.edu [BejeranoFall13/14]
incompatible
29
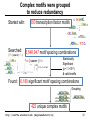
Complex motifs were grouped
to reduce redundancy
Started with:
300 transcription factor motifs
…
Searched:
(TF1 {spacer} TF2)
6,548,947 motif spacing combinations
Fox { spacer } Ets
300
50 2 6.5mil
2
Statistically
Significant
(p < 1×10-8)
& valid motifs
Found: 6,180 significant motif spacing combinations
Grouping
422 unique complex motifs
http://cs273a.stanford.edu [BejeranoFall13/14]
30
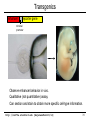
Transgenics
enhancer
reporter gene
minimal
promoter
Observe enhancer behavior in vivo.
Qualitative (not quantitative) assay.
Can section and stain to obtain more specific cell-type information.
http://cs273a.stanford.edu [BejeranoFall13/14]
31
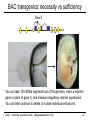
BAC transgenics: necessity vs sufficiency
You can take 100-200kb segments out of the genome, insert a reporter
gene in place of gene X, and measure regulatory domain expression.
You can then continue to delete or mutate individual enhancers.
http://cs273a.stanford.edu [BejeranoFall13/14]
32
Gene Regulation: Enhancers are modular and additive
limb
neural
tube
brain
Sall1
Temporal gene expression pattern “equals”
sum of promoter and enhancers expression patterns.
http://cs273a.stanford.edu [BejeranoFall13/14]
33
Genome Engineering
Technologies are in
fact constantly
improving that allow
us to edit the nuclear
genome itself.
Edit the genome of an
embryonic stem cell,
breed homozygous
modified animals.
http://cs273a.stanford.edu [BejeranoFall13/14]
34
Chromosome conformation capture (3C)
People are also developing methods to detect
when two genomic regions far in sequence
are in fact interacting in space.
Ultimately this will allow to determine
experimentally the regulatory domain of
each gene (likely condition dependent).
http://cs273a.stanford.edu [BejeranoFall13/14]
35
4C example result (in a single context)
TSS probe
Irreproducible peaks
http://cs273a.stanford.edu [BejeranoFall13/14]
36
Gene Regulation is HOT
Despite its complexity gene regulation is currently one of
the hottest topics in the study of the human genome.
Large projects are pouring tons of money to generate huge
descriptive datasets.
The challenge now is to glean logic from these piles.
To be continued…
http://cs273a.stanford.edu [BejeranoFall13/14]
37