* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download The initiation phase of protein synthesis in eukaryotes
Peptide synthesis wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Western blot wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Lipid signaling wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Citric acid cycle wikipedia , lookup
Signal transduction wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Paracrine signalling wikipedia , lookup
Biochemical cascade wikipedia , lookup
Ligand binding assay wikipedia , lookup
Polyadenylation wikipedia , lookup
Biochemistry wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Two-hybrid screening wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Proteolysis wikipedia , lookup
Gene expression wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Transfer RNA wikipedia , lookup
Biosynthesis wikipedia , lookup
The initiation phase of protein synthesis in eukaryotes
The final phase of protein
synthesis - Translational
Termination
The structure of a human translation release factor (eRF1) and its
resemblance to a tRNA molecule
Translational Control
Two Types of Translational Control
Two Types of Translational Control
1] Global regulation
Two Types of Translational Control
1] Global regulation
2] Message specific regulation
Review of Eukaryotic mRNA translation
• Initiation -
Recruitment of the ribosome to the mRNA
Recognition of the start codon
• Elongation -Movement of the ribosome along the mRNA
Decoding the mRNA into an extending polypeptide chain
• Termination - Recognition of the stop codon
Release of the ribosome and protein chain from the mRNA
start
stop
7mGpppN
5’UTR
ORF
3’UTR
AAAAAAAAAAA
Review of Eukaryotic mRNA translation
• Initiation -
Recruitment of the ribosome to the mRNA
Recognition of the start codon
start
stop
7mGpppN
5’UTR
ORF
3’UTR
AAAAAAAAAAA
Initiation
Recruitment of ribosome and the initiator methionyl tRNA to the start codon
Initiation
Recruitment of ribosome and the initiator methionyl tRNA to the start codon
eIF1A
eIF2
eIF2B
eIF3
eIF4A
eIF4B
eIF4E
eIF4G
eIF5
eIF5B
aminoacylated initiator methionyl tRNA (Met-tRNAiMet)
40S ribosomal subunit
60S ribosomal subunit
GTP
ATP
Initiation
Recruitment of ribosome and the initiator methionyl tRNA to the start codon
eIF1A
eIF2
eIF2B
eIF3
eIF4A
eIF4B
eIF4E
eIF4G
eIF5
eIF5B
aminoacylated initiator methionyl tRNA (Met-tRNAiMet)
40S ribosomal subunit
60S ribosomal subunit
GTP
ATP
Five Steps1. 40S ribosomal subunit and tRNAiMet
preparation
2.
mRNA selection and preparation
3.
40S/ mRNA binding
4.
scanning and AUG recognition
5.
60S ribosomal subunit joining
Step 1: 40S ribosomal subunit and tRNAiMet preparation
60S
60S
3
40S
eIF2•GTP•Met-tRNAiMet is called the Met
Ternary Complex-TC
3
2
GTP
40S
1A
1A
1
Met
Met
Met-tRNAiMet
2
GDP
GTP
2B
GTP
2
GDP
3
1
2
GTP
1A
43S pre-initiation complex
Formation of Ternary Complex an important regulatory node
60S
60S
3
40S
eIF2•GTP•Met-tRNAiMet is called the Met
Ternary Complex-TC
3
2
GTP
40S
1A
1A
1
Met
Met
Met-tRNAiMet
2
GDP
GTP
2B
GTP
2
GDP
3
1
2
GTP
1A
43S pre-initiation complex
Step 2: mRNA selection and preparation
•
The mRNA is bound by eIF4F complex composed of eIF4E, eIF4G, eIF4A.
•
eIF4B binds to eIF4A and facilitates the helicase activity of 4A that is required to
unwind secondary structures in the mRNA during scanning
eIF4F
4E
Cap
4G
4B
4A
AUG
11
Step 2: mRNA selection and preparation
eIF4E binding is an important
regulatory node
4E
Cap
eIF4F
4G
4B
4A
AUG
12
Step 3: 43S/ mRNA binding and scanning
43S mRNA interaction
eIF3 in the 40S complex and eIF4G in the mRNA complex interact
Met
3
1
2
GTP
1A
43S pre-initiation complex
4G
4E
4B 4A
Cap
3
AUG
48S pre-initiation complex
4E
4G
Cap
3
4B 4A
Met
1
GTP
2
1A
AUG
Step 3: 43S/ mRNA binding and scanning
Scanning
• The 48S complex scans each codon in a 5’ to 3’ direction looking for an AUG.
• The eIF4A helicase activity irons out RNA hairpins allowing the 40S complex to move.
• multiple rounds of ATP hydrolysis are required to provide energy for the movement
4E
Cap
4G
4B 4A
GTP
3 Met
2 1A
1
AUG
Step 4: AUG recognition and GTP hydrolysis
AUG recognition
• eIFs 1, 2 and 5 are required for this step.
5
Met
3
Cap
2
5
1
1A
AUG
GTP
GTPase step and recycling of eIFs
• eIF5 stimulates the GTPase activity of eIF2 (GTP hydrolysis to GDP)
leading to loss of initiation factor binding.
Met
1A
Cap
AUG
2
GDP
3
1
5
Step 5: 60S Joining
A second G protein eIF5B promotes 60S subunit joining. Hydrolysis
of GTP to GDP promotes 5B and eIF1A release. The elongation
phase, where the protein is synthesized can now begin.
60S
5B
GTP
Met
Cap
AUG
1A
GTP
GDP
Met
AUG
Cap
5B
GDP
1A
16
Elongati
on
Measurement of Translation
Reporter Assays
Reporter
luciferase
B-gal
GFP
poly(A)
Westerns
Extract proteins
Run on SDS-Page
Probe with Antibody
35S incorporation
Pulse-label 35S met or cys
Take time points
CPM incorporated (X103 )
5
4
3
2
1
0
Measure incorporation by TCA prec.
followed by scintillation
0
2
4
6
Time (mins)
8
10
Yeast Extract
Polysome analysis
Sucrose
gradient
Non-translating
RNAs
mRNA
80S
Translation efficiency
260nm
Spin
Abs
polyribosomes
Collect fractions
60S
mRNA
40S
40S subunit
Top
1 2
60S subunit
Fractions
3
4
5
6
7
8
Bottom
9 10 11 12 13 14 15 16
polyribosomes
mRNAs
80S ribosome
rRNA
28S rRNA
18S rRNA
Efficiently translated
mRNA
Poorly translated
mRNA
Two Types of Translational Control
1] Global regulation
2] Message specific regulation
1] Global regulation
80S
RNP
P
0 min
Condition A
120 min
Condition B
GAL-Dhh1p
1] Global regulation
80S
RNP
P
0 min
Condition A
120 min
Condition B
GAL-Dhh1p
Usually seen during conditions of
stress or stimulation by hormone
1] Global regulation
eIF-4E regulation
Ternary Complex formation
Step 3: 43S/ mRNA binding and scanning
43S mRNA interaction
eIF3 in the 40S complex and eIF4G in the mRNA complex interact
Met
3
1
2
GTP
1A
43S pre-initiation complex
4G
4E
4B 4A
Cap
3
AUG
48S pre-initiation complex
4E
4G
Cap
3
4B 4A
Met
1
GTP
2
1A
AUG
Three Types of eIF4E regulation
1] Transcriptional
2] eIF4E phosphorylation
3]Binding of inhibitory proteins
Transcriptional Regulation of eIF4E
eIF4E is the least abundant initiation factor in cells
Transcription increase several fold in fibroblast upon growth factor treatment
Mechanism poorly understood, but involves c-Myc regulation.
Links to cancer
Phosphorylation of eIF4E
eIF4E phosphorylated at a single site -Ser209
Phosphorylation increases translation rate
Structure of eIF4E with m7GpppG analog
Ser209
m7GpppG
Phosphorylated Ser209 interacts with Lys159 to make a retractable salt bridge
Clamping mRNA in the cap-binding slot
Ser209
Lys159
12/2/99
932
Growth Factor, Hormones, Mitogens,8505_AR_28
and Cytokines
12/2/99 6:44 mediate
PM Page 933 eIF4E
phosphorylation
Page 932
eIF4 INITIATION FACTORS
GINGRAS, RAUGHT & SONENBERG
TABLE 1
TABLE 1
Effect of various stimuli on eIF4E and 4E-BP1 phosphorylation
Stimulus
Cell type
Adenovirus
infection (early)
Stimulus
Cell type
+b
227, 228
Insulin-like
growth factor I
Rat aortic
smooth muscle
+
371
+
208, 357
Swiss 3T3-L1
+
362, 367
+
235
Insulin-like
growth factor II
358
Interleukin 1!
CHO.K1
Interleukin 3
Myeloid progenitor
Lipopolysaccharide
B lymphocytes
+
212
L-Pyrroline-5carboxylic acid
Rabbit reticulocyte
lysate
+
372
Nerve growth factor
PC12
+
Platelet-derived
growth factor
NIH 3T3
Lung fibroblasts
Swiss 3T3-L1;
aortic SM
+
+
4E-BP1
HeLa, 293
-a
Angiotensin II
Vascular smooth muscle
+
Anisomycin
Swiss 3T3, 293
CD4+
or
Mature
thymocytes
CD8+
+
Arsenite
293, CHO.K1
+
Concanavalin A
Peripheral blood mononuclear cells
+
DAMGOc
CHO overexpressing
"-receptor
Epidermal growth
factor
Mammary
epithelial cells
P19
!
177
359
+
360
+
274
+
361
Swiss 3T3, 3T3-L1,
L1, PC12
+
Gastrin
AR4-2J tumor cells
+
258
GMCSF + SLF
Hematopoietic MO7e
+
365
Heat shock
Reuber hepatoma
+
231
High glucose
Isolated rat pancreatic
islets
+
NIH 3T3
+
3T3-L1
+
Insulin
+
362–364
366
178, 193, 319
+
162, 362, 363
367
CHO
+
199, 211
Skeletal muscle
+
368, 369
Swiss 3T3; 32D;
293; CHO-IR
+
227, 235, 238
370
(continued)
Gingras, unpublished data). Nevertheless, these data argue that ERKs and p38
act as upstream effectors of eIF4E phosphorylation (Figure 5). It is unclear why
eIF4E phosphorylation is not increased after treatment of cells with sorbitol,
H2O2, or heat shock, which are potent activators of p38 MAP kinases (177).
One hypothesis is that these stresses also decrease phosphorylation of the eIF4E-
933
(continued)
References
eIF4E
Anti-CD3
Annu. Rev. Biochem. 1999.68:913-963. Downloaded from arjournals.annualreviews.org
by CASE WESTERN RESERVE UNIVERSITY on 02/22/06. For personal use only.
6:44 PM
Annu. Rev. Biochem. 1999.68:913-963. Downloaded from arjournals.annualreviews.org
by CASE WESTERN RESERVE UNIVERSITY on 02/22/06. For personal use only.
8505_AR_28
eIF4E
4E-BP1
+
References
177
+
238
+
194, 364
+
178, 193, 319
373
362, 367, 371
PHA + phorbol ester Human T cells
+
170
Phorbol ester
NIH 3T3, CHO,
PBL, B cells
+
178, 193, 199,
211, 212, 319, 359
3T3-L1,
leukaemic T cells,
retic. lysate
Swiss 3T3
+
162, 359, 372
NIH 3T3
Swiss 3T3
CHO
3T3-L1
+
+
+
U937, HeLa,
ME180, BAEC, FS,
HUVEC
+
Serum
Tumor necrosis
factor "
+
235
+
178, 193, 256, 319
374
211
362, 367
+
177, 375
a-,
No change in phosphorylation
Increase in phosphorylation; blank space indicates not determined
cAbbreviations: DAMGO, [D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin; GMCSF, granulocyte-macrophage colony stimulating
factor; SLF, steel factor; PHA, phytohemagglutinin.
b+,
An excellent candidate for the eIF4E kinase is the MAP kinase-interacting
protein kinase-1(MNK1; also called MAP kinase signal-integrating kinase).
MNK1 was identified independently by two groups as a substrate for ERK1 and
8505_AR_28
12/3/99
12:50 PM
Page 935
Signal Transduction Pathways Mediate eIF4E Phosphorylation
99.68:913-963. Downloaded from arjournals.annualreviews.org
RESERVE UNIVERSITY on 02/22/06. For personal use only.
eIF4 INITIATION FACTORS
935
Figure 5 Signaling pathways to eIF4E phosphorylation. The intracellular signaling
pathways leading to eIF4E phosphorylation are diagrammed.
AlsoMAP
indicated
are the
MNK1=
kinase-interacting
protein kinase
inhibitors (in italics) used to delineate the pathways. Both MNK1 and eIF4E interact
with eIF4G, bringing the two proteins in close proximity, resulting in more efficient
eIF4E phosphorylation.
eIF4E interacting proteins
eIF4E binding proteins exist in higher eukaryotes (4E-BP)
Three found in mammals (4E-BP1, 4E-BP2, and 4E-BP3)
Small (10-12 kDa), heat-stable, all contain eIF4E binding site
Binding of 4E-BP does not alter eIF-4E affinity for the cap
et aL
4E-BP
proteins
block eIF4E/eIF4G binding
A.Haghighat
-
B
--I,
+
H A-plY2n)
i ;jS T
-*
,43i
P
i
J1-F F
1
Fig. 3. 4E-BP1 and p220 compete for binding to eIF-4E. (A) m7GDP-coupled agarose resin was inc
ml) and
recombinant murine eIF-4E in buffer A. The resin was washed in buffer A (3XHaghighat
et al., then
1995 incub
for 60 min at 4° C. The resin was washed in buffer A (3x 1 ml) and incubated further with buffer A
Note: p220 old
nomenclature for eIF4G
4E-BP proteins block eIF4E/eIF4G binding
eIF4G peptide
4E-BP peptide
GINGRAS, RAUGHT & SONENBERG
Annu. Rev. Biochem. 1999.68:913-963. Downloaded from arjournals.annualreviews.org
by CASE WESTERN RESERVE UNIVERSITY on 02/22/06. For personal use only.
938
Figure 6 The cocrystal structure of the 4E-BP1 and eIF4GII peptides bound to
eIF4E. (A) The human 4E-BP1 peptide (orange) and the m7GDP cap analog (green)
bind to opposing regions of eIF4E (blue); (B) The 4E-BP1 peptide binds to the dorsal
Gingras
et al., 1999
convex surface of eIF4E and adopts an extended L-shaped conformation; (C)
the
eIF4GII peptide (red) also adopts an extended L-shaped conformation when binding
8505_AR_28
12/3/99
1:32 PM
Page 940
4E-BP binding to eIF4E is regulated by phosphorylation
ownloaded from arjournals.annualreviews.org
ERSITY on 02/22/06. For personal use only.
940
GINGRAS, RAUGHT & SONENBERG
Figure 7 The binding of the 4E-BPs to eIF4E is regulated by phosphorylation. The
4E-BPs and eIF4Gs compete for a common binding site on eIF4E. Various stimuli increase the phosphorylation of the 4E-BPs. Hyperphosphorylated 4E-BPs have a relatively low affinity for eIF4E. Conversely, a decrease in 4E-BP phosphorylation increases the affinity of the 4E-BPs for eIF4E. Free eIF4E interacts with eIF4G to form
the translationally active eIF4F complex. GPCR: G protein coupled receptor.
Activation of eIF4E by two distinct yet re-enforcing
mechanisms
!"#$%&'()&*!&
+,-.+,-/01(2!3 34/!'210&*0&
5)678&9,4',&4.&3-'6!3&-)&
!"#$:&; 4)'/!(.!.&(<<4)420&
<-/&2,!&=>&'(+&(*-?2&$@
!"#$%&4.&(1.-&('24A(2!3&*0&
+,-.+,-/01(24-)&-<&42.&
*4)34)B&+/-2!4)&2,/-?B,&
'!11&B/-92,&.4B)(14)B
!
1] Global regulation
eIF-4E regulation
Ternary Complex formation
Step 1: 40S ribosomal subunit and tRNAiMet preparation
60S
60S
3
40S
eIF2•GTP•Met-tRNAiMet is called the Met
Ternary Complex-TC
3
2
GTP
40S
1A
1A
1
Met
Met
Met-tRNAiMet
2
GDP
GTP
2B
GTP
2
GDP
3
1
2
GTP
1A
43S pre-initiation complex
Amino acid depletion induces a rapid translational repression events in yeast
80S
RNP
P
0 min
120 min
-Amino
GAL-Dhh1p
+Amino
Acids
Acids
Despite this, some mRNA are upregulated
Alan Hinnebusch and Colleagues
5.59:407-450. Downloaded from arjournals.annualreviews.org
ESERVE UNIVERSITY on 02/22/06. For personal use only.
ANRV253-MI59-18
ARI
4 August 2005
16:40
The Presence of uncharged tRNAs stimulates eIF2α phosphorylation
Ternary Complex Levels High
Non-starvation
conditions
Ternary Complex Levels Low
Starvation conditions
Figure 3
Model for GCN4 translational control. GCN4 mRNA and reinitiating ribosomes are depicted as in
Step 1: 40S ribosomal subunit and tRNAiMet preparation
60S
60S
3
40S
eIF2•GTP•Met-tRNAiMet is called the Met
Ternary Complex-TC
3
2
GTP
40S
1A
1A
1
Met
Met
Met-tRNAiMet
2
GDP
GTP
2B
GTP
2
GDP
3
1
2
GTP
1A
43S pre-initiation complex
NTD is sufficient for binding to a C-terminal
segment of Gcn1p (approximately residues
2050–2400), and overexpressing either domain dissociates the native Gcn1p/Gcn2p
complex and produces a Gcn− phenotype that
Gcn1p function without affecting ribosome
binding or Gcn20p binding by Gcn1p, thus
showing that the Gcn1p-Gcn2p interaction
is crucial. The extreme N-terminal (amino
acids 1–672) and C-terminal (amino acids
Annu. Rev. Microbiol. 2005.59:407-450. Downloaded from arjournals.annualreviews.org
by CASE WESTERN RESERVE UNIVERSITY on 02/22/06. For personal use only.
GCN2 kinase is a sensor for uncharged tRNA
422
Hinnebusch
Amino acid depletion induces a rapid translational repression events in yeast
80S
RNP
P
0 min
120 min
-Amino
GAL-Dhh1p
+Amino
Acids
Acids
Despite this, some mRNA are upregulated
Alan Hinnebusch and Colleagues
!"#$%&'()*+#%+,%-%.'-#,"'+/)+(#%0-")('%)1-)%
2*,/(#3,%)(%45+#(%4"+3%6*7+"+*#"8
"
#!%
#
9
!
9
$
!"#$
& :8#)1*,+,%(7%!"#$/%+,%'*;<=-)*3%-)%)1*%)'-#,=-)+(#-=%=*>*=
& ?)%+,%'*;<=-)*3%@8%-#%*?0A! B+#-,*C%!"#A/
& 4")+>+)8%(7%!"#A/%+,%"(#)'(==*3%@8%<#"1-';*3%)2D4
icrobiol. 2005.59:407-450. Downloaded from arjournals.annualreviews.org
WESTERN RESERVE UNIVERSITY on 02/22/06. For personal use only.
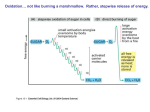
Figure 3
Model for GCN4 translational control. GCN4 mRNA and reinitiating ribosomes are depicted as in
Figure 2. The three subunits of eIF2 and the five subunits of eIF2B are listed in the left panel. Negative
regulatory factors are depicted in red; positive effectors are depicted in green. Following translation of
uORF1, ∼50% of the 40S ribosomes remain attached to the mRNA and resume scanning. Under
nonstarvation conditions, the 40S subunit quickly rebinds the TC and reinitiates at uORF4 because the
The GAAC response a translation control pathway that allows for survival when times
are tough
Amino acid levels reduced
uncharged tRNA levels high
Exogenous amino acid levels high
charged tRNA levels high
GCN2 non-ribosome bound, inactive
eIF2α unphosphorylated
GCN2 ribosome bound, active
eIF2α phosphorylated
Ternary complex levels reduced
Ternary complex levels high
Bulk translation decreases
Translation rate high
GCN4 mRNA translated
Gcn4p dependent transcription of
amino acid biosynthesis genes occurs
!"#$%&#'(')*+! ,%-./'/ %-(0"1.234$'/
" 567+(#$%&'()$&*+&,'-,&(.$/*01*('',2(,&(.$3
" 8,9(#2.%45)6+&',$2)2*7893
" 809,(#:7*+&')++/*%$;.52)2*<'.&)($*')+<.$+)3
" :9)(#=)>)/*')&(?%5.?@&)+3
9';"#.$%4-(4<(
$2.-/=2%&$%4-(<.=$42(
>?*@(A3(')*+!
1%-./'/ %-(
B.BB.#%.-(='##/
5>DDE@
8086,
6:C8
F$2'//(.=$%G.$'H(;'-'/
A,'2($B*,$2*7.$
!"#$%&'()*&!"#$%&+,-&./0#-&1*2.(1*3&451&
61,7-',6(57,'&857615'&45''59(7:&-61*-''''
!"#$%
!"#$&
!"#$%&'( *!+,-.
'($)
!"#$%&'()& &*+'&))$,-./*.01234
5
65
%,78.$-8"#9$,-
&
/
0
%11
%11
%
%&11
%111
123
%%11
411
123
#56.-789:6;7-<=>?6
@#5*-::-<,.;6A#6
+=7B++
;56+&./0#-&,1*&1*2.(1*3&451&61,7-',6(57,'&
857615'&54&!"#$&<=&>0&-61*-"#$%&'()#'%)*+'
Two Types of Translational Control
1] Global regulation
2] Message specific regulation