* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Translation
Molecular cloning wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Metagenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
History of genetic engineering wikipedia , lookup
Messenger RNA wikipedia , lookup
Microevolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
History of RNA biology wikipedia , lookup
Transfer RNA wikipedia , lookup
Frameshift mutation wikipedia , lookup
Epitranscriptome wikipedia , lookup
Non-coding RNA wikipedia , lookup
Primary transcript wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Expanded genetic code wikipedia , lookup
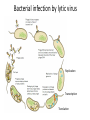
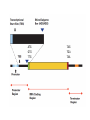
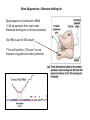
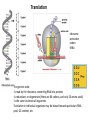
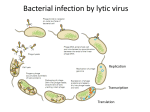
Bacterial infection by lytic virus Replication Transcription Translation Central Dogma of Molecular Biology replication DNA transcription RNA translation Protein Phage genes are replicated/transcribed/translated by bacterial machinery Replication – DNA Polymerase Transcription- RNA polymerase Translation- Ribosome “Central Dogma of Molecular Biology” Flow of information from DNA RNA Protein Schematic image of gene structure ATG GTG TTG TAG TGA TAA ATG GTG TTG TAG TGA TAA Shine Dalgarno box = Ribosome binding site Signal sequence in prokaryotic mRNA ~4-14 bp upstream from start codon Ribosome binding site to initiate translation 16s rRNA is part of 30S subunit **You will look for a “SD score” as one measure of a good start codon prediction. Translation ribosome anticodon codon tRNA The genetic code: -Is read by the ribosome, converting RNA into proteins -Is redundant, or degenerate (there are 64 codons, and only 20 amino acids) -Is the same in almost all organisms Translation in individual organisms may be biased towards particular tRNA pool, GC content, etc. The Ribosome recognizes the correct reading frame Silly example: the mad cat ate the fat rat and the big bat t hem adc ata tet hef atr ata ndt heb igb at th ema dca tat eth efa tra tan dth ebi gba t Science example: ATGAATATGAAAACAACTATTAAAGCAGGTCAATTCGAATTAGGACAAGTG Coding Potential Organisms have a preference for certain codons. Six frame translation This document shows the six potential reading frames, with each predicted gene highlighted. protein1 protein2 Frame 1 Frame 2 Frame 3 Frame 4 Frame 5 Frame 6 Coding Potential Organisms have a preference for certain codons. This plot shows predicted open reading frames based on ‘coding potential’. Frame 1 Frame 2 Frame 3 What does coding potential tell you? Upticks are start codons Downticks are stop codons Predicts open reading frames (thick black bar) based on how well the codon usage matches 1) The host bacteria (black line) 2) The phage (red line) BLAST: Basic Local Alignment Search Tool • • • • The most highly used bioinformatics tool? Finds similarity between biological sequences (DNA or Protein) Compares your favorite protein to the “non-redundant” database Shows alignment and calculates statistical significance BLASTn uses nucleotide input and compares to a nucleotide sequence database BLASTp uses amino acid input and compares to a proteni sequence database Evaluating a BLASTp match Query- your protein sequence Subject- the match from the database Look through alignment, what kinds of scores are represented? Look through text list of hits, how does your Query coverage and E value look? How does % identity look? Do the words describing the hits tell you anything? Look at the first few alignments: Query and Subject are aligned. Identical amino acids are shown by the same letter being in the middle of the two sequences. + means the amino acids are similar. A blank means they are different. – means the computer put in a ‘gap’ (not real) so it could make the best alignment possible. ***Do you get a 1:1 match of query:subject? Ribosomes recognize the Shine-Dalgarno sequence “AGGAGG” core 4-14 nucleotides upstream of start DNA Frames view gives you the “SD score” Work through the annotation of gp1 using the guiding principles of annotation Start/stop coordinates Enter after you make your choice Length In features table or description tab Longest ORF? Yes, or no but… Gap/overlap to previous gene Can ignore for gp1, remember gaps -1, overlaps +1 Predicted by Genemark or Glimmer or Self SD Score In frames view, is it the highest? Coding potential In GeneMark files on wiki Best BLASTp match Is it 1:1 and for most of the query length? Function Based on homology or ‘Hypothetical Protein’ Function source Blastp conserved domain, Phamerator, hhpred Homework: Annotate the first three genes! -post your annotations on your wiki page **every semester one or two students lose their annotations -be sure to save your DNAMaster file on your computer in Documents and somewhere else (USB or wiki)