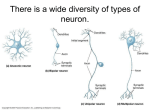
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Cell Ontology – INCF Neuron Workshop
Survey
Document related concepts
Patch clamp wikipedia , lookup
Signal transduction wikipedia , lookup
Nonsynaptic plasticity wikipedia , lookup
Feature detection (nervous system) wikipedia , lookup
Neurotransmitter wikipedia , lookup
Subventricular zone wikipedia , lookup
Molecular neuroscience wikipedia , lookup
Neuroanatomy wikipedia , lookup
Synaptic gating wikipedia , lookup
Single-unit recording wikipedia , lookup
Stimulus (physiology) wikipedia , lookup
Electrophysiology wikipedia , lookup
Nervous system network models wikipedia , lookup
Transcript
Cell Ontology – INCF Neuron Workshop April 12-14, 2011 Loudermilk Conference Center, Atlanta Georgia Supported by NHGRI HG002273-09Z and The International Neuroinformatics Coordinating Facility Co-sponsored by the Georgia State University Center for Neuromics Thanks to • • • • • • • Janis Breeze, INCF Jyl Boline, INCF Paul Katz, GSU Alan Ruttenburg, SUNY Buffalo Judith Blake, The Jackson Laboratory Ashley Stanton, The Jackson Laboratory Beth Partridge, Hurley Travel at Jax Participants Giorgio Ascoli, George Mason University Judith Blake, The Jackson Laboratory Mihail Bota, University of Southern California (via teleconference) Janis Breeze, INCF Program Officer Robert Burgess, The Jackson Laboratory Jyl Boline, INCF Project Coordinator (via teleconference) Alexander Diehl, SUNY Buffalo Akshaye Dhawan, Ursinus College Chad Frederick, Georgia State University Daniel Gardner, Weil Medical College, Cornell Melissa Haendel, Oregon Health & Science University David Hamilton, George Mason University Sean Hill, INCF Excecutive Director Kei Ito, The University of Tokyo Saurav Karmakar, Georgia State University Paul Katz, Georgia State University Stephan Larson, University of California, San Diego Ed Lein, Allen Brain Institute Terrence Meehan, The Jackson Laboratory Jose Leonardo Mejino, University of Washington Chris Mungall, Lawrence Berkeley National Laboratory Sasha Nelson, Brandeis University Sridevi Polavaram, George Mason University David Osumi-Sutherland, Cambridge University (via teleconference) Abir Ashfakur Rahman, Georgia State University Patrick Ray, SUNY Buffalo Kathleen Rockland, Massachusetts Institute of Technology Alan Ruttenburg, SUNY Buffalo Deboleena Roy, Emory University Joseph Shea, SUNY Buffalo Gordon Shepherd, Yale University Raj Sunderraman, Georgia State University Menno Witter, Norwegian University of Science and Technology Workshop Goals • Introduction to the Cell Ontology • Review of neuron ontology efforts • Discussion of the ontological representation of neurons • Review of core set of high level neuron terms • Set up Working Groups • Working Groups develop neuron terms and report back Satellite Meeting • For in depth discussion of ontology development-related issues • Discussion points to be collects during main meeting • For instance, “How do we present lines of evidence for asserted neuron properties as part of the ontology?” The Cell Ontology • An ontology of cell types built by biologists for the needs of data annotation and analysis. • The Cell Ontology covers in vivo cell types from all of biology. • The Cell Ontology is not – A list of specific cell lines, immortal or otherwise, although the Cell Ontology may be used to describe such cells if they correspond to an in vivo cell type. – A list of in vitro methods for preparing cell cultures, although the Cell Ontology may be used to describe the resulting cells if they correspond to an in vivo cell type. OBO Reference Ontologies RELATION TO TIME CONTINUANT INDEPENDENT OCCURRENT DEPENDENT GRANULARITY ORGAN AND ORGANISM Organism (NCBI Taxonomy?) CELL AND CELLULAR COMPONENT Cell (CL) MOLECULE Anatomical Organ Entity Function (FMA, (FMP, CPRO) Phenotypic CARO) Quality (PaTO) Cellular Cellular Component Function (FMA, GO) (GO) Molecule (ChEBI, SO, RnaO, PrO) Molecular Function (GO) Organism-Level Process (GO) Cellular Process (GO) Molecular Process (GO) Smith et al., Nature Biotechnology 25, 1251 - 1255 (2007) Overview of Cell Ontology • Developed by Jonathan Bard, David States, Michael Ashburner, and Seung Rhee and first described in 2005, Genome Biology, 6:R21. • The Cell Ontology is sometimes called the Cell Type Ontology, and often referred to as the “CL”, based on its identifier space. For instance the term “B cell” has the ID “CL:0000236” • The CL currently has over 1500 cell type terms, over 500 of which have logical definitions. Textual Definitions • • • • name: CD4-positive, CD25-positive, alpha-beta regulatory T cell def: "A CD4-positive, CD25-positive, alpha-beta T cell that regulates overall immune responses as well as the responses of other T cell subsets through direct cell-cell contact and cytokine release.” name: induced T-regulatory cell def: "CD4-positive alpha-beta T cell with the phenotype CD25-positive, CTLA-4-positive, and FoxP3-positive with regulatory function." Logical Definitions Logical Definitions Pre-reasoning Post-reasoning Logical Definitions of Neurons • Retinal Ganglion Cell – def: "A neuron located in the ganglion cell layer of the eye that receives inputs via bipolar, horizontal and amacrine cells. The axons of these cells make up the optic nerve." [GOC:add, GOC:dph] CL Neuron Improvements so far • We have added textual definitions for around 120 of the existing neuron types in CL, and logical definitions for around 75 of these. • With help from INCF, we have imported 130 neuron types from the BAMS (Brain Architecture Management System) and created new genusdifferentia textual definitions for these. About 40 of these have been given logical definitions. • These changes are still under review. Neurons are complicated • Most neurons are defined in part by their anatomical location. – Brain and nervous system anatomy varies considerably between species. – Many anatomical systems exist for brains of different species. These are often in conflict with each other. – Neurons often have their soma in one anatomical location, axon in another, and dendrites in a third. • We hope upper level neuron classes that can be defined reliably across species. – – – – Morphology Neurotransmitter secretion Large scale anatomical structures Function What does it mean to “represent” a Neuron in an Ontology? • Capture basic properties of particular neurons in a textual definition based on the literature. The combination of properties should uniquely define the neuron. • Translate these properties to a logical definition (cross-product), based on relations that link the neuron to terms in other ontologies. • Not a mathematical model. Neuron Ontology and Database Efforts • • • • • • • • Neurolex and NIF-cell INCF Neuron Registry Task Force Petilla Interneuron Nomenclature Group SenseLab NeuronBank Brain Architecture Management System (BAMS) Foundational Model of Anatomy Virtual FlyBrain