* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download medmicro4-weapons delivery – G+
Model lipid bilayer wikipedia , lookup
Theories of general anaesthetic action wikipedia , lookup
Cytokinesis wikipedia , lookup
Magnesium transporter wikipedia , lookup
SNARE (protein) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Cell membrane wikipedia , lookup
Type three secretion system wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Endomembrane system wikipedia , lookup
List of types of proteins wikipedia , lookup
Signal transduction wikipedia , lookup
Western blot wikipedia , lookup
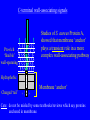
SBM 2044: Lecture 4 Weapons delivery & deployment (Part II) Secretion & targeting of protein virulence factors in Gram-positive bacteria Sec-dependant General secretion pathway (GSP) Gram-negative bacteria Proteins reach periplasm, but OM is additional barrier need other mechanisms to get protein out thro’ OM. (Types I - V secretion) Gram-positive bacteria Sufficient to get protein out. In this case, other mechanisms needed to retain wall - associated proteins OM IM Type II secretion sec sec Signal-peptide Targeting secreted proteins to Gram-positive cell walls Four distinct mechanisms identified to date: Rare: • Binding to wall teichoic acid • Binding to membrane anchored LTA More widespread: • Lipoprotein ‘anchors’ • C-terminal wall-associating signals 1. Binding to cell-wall teichoic acid Streptococcus pneumoniae and Streptococcus suis Pneumococcal surface protein A (PspA) Pneumococcal autolysin (LytA) S. suis autolysin- [homologous to pneumococcal LytA] C-terminal ends share homologous choline-binding domains – enable binding to TA of these species The structure of teichoic acid: Polymer of either Glycerol phosphate or Ribitol phosphate, with various substituents (R) poly-ribitol phosphate O O P O O H H H H H C C C C C H O OH O H R O O P O R’ H O C H n In most species studied to date R = D-alanine R’ = N-acetylglucosamine In S. pneumoniae and S. suis R = phosphodiester linked choline - chemically more stable than ester-linked D-Ala 2. Binding to membrane anchored LTA Single example recognised only recently - InlB of Listeria monocytogenes – has C-terminal domain that ‘targets’ LTA – mechanism?? 3. Lipoproteins • attached at outer surface of cytoplasmic membrane by a lipid anchor Examples include penicillinase in S. aureus • Similar mechanisms used in both Gram + & Gram . Distinctive N-terminal signal peptides recognized by distinct Sec apparatus with specialized signal peptidase (called signal peptidase II) Lipoprotein signal peptides NShort hydrophobic 1-3 positively sequence charged a.a. Signal peptidase II cleavage site -Leu-x-y- Cysx and y usually small, uncharged residues A diglyceride is attached to the N-terminal Cys of the mature protein Diglyceride Contrast with ‘typical’ GSP secretion signal-peptide ( Lecture 3 ) 4. ‘Sorting’ via C-terminal wall-associating signals Vast majority of Gram + wall-associated proteins share structurally similar C-terminal wall-associating signals Hydrophobic /Charged ‘tail’ membrane ‘anchor’ -C Pro-rich region LPxTG motif 15 - 20 hydrophobic residues 5 - 10 mostly charged C-terminal wall-associating signals Studies of S. aureus Protein A, showed that membrane ‘anchor’ plays a transient role in a more complex wall-associating pathway Pro-rich ‘flexible’ wall-spanning Hydrophobic Charged ‘tail’ Membrane ‘anchor’ + + Care: do not be misled by some textbooks/reviews which say proteins anchored in membrane. C N-terminal signal peptide N Wall-associating signal Signal peptidase wall-associated ‘Sortase’ Cleavage at LPxTG N mRNA G Cross-linked to cell-wall Some, but not C Majority Minority necessarily all, ‘cleaved’ simply covalently ‘anchored’? at LPxTG linked to wall (e.g. ActA in Listeria) (e.g. InaA, Prot. A) Retaining secreted proteins in Gram-positive cell walls 1. Binding to wall teichoic acid Limited to a very few species (e.g. S. pneumoniae, S. suis) 2. Binding to membrane anchored LTA Single example recognised only recently (InlB of Listeria monocytogenes) 3. Lipoprotein ‘anchors’ A minority of wall-associated proteins in many species anchored to outer surface of cell membrane via an N-terminal lipid anchor 4. C-terminal wall-associating signals Vast majority of wall-associated proteins studied to date share structurally similar C-terminal wall-associating signals Retaining proteins at Gram-negative cell-surfaces First step: Sec-dependent secretion to periplasm (GSP) Then: • Targeting of integral OM proteins - OM-interacting ‘surfaces’ result from folding in periplasm (may involve periplasmic Dsb and Ppi enzymes) OR • Individual biogenesis pathways – e.g. fimbriae References • Navarre and Schneewind. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope (1999). Microbiology and Molecular Biology Reviews, 63, 174-229. • Ton-That et al. Protein sorting to the cell wall envelope of Gram-positive bacteria (2004). Biochimica et Biophysica Acta, 1694, 269-278.