* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Tmm - OpenWetWare
Long non-coding RNA wikipedia , lookup
Transposable element wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Human genome wikipedia , lookup
Copy-number variation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Oncogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Pathogenomics wikipedia , lookup
Public health genomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Ridge (biology) wikipedia , lookup
The Selfish Gene wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression programming wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
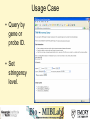
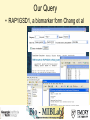
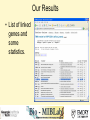
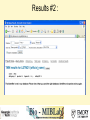
Tmm: Analysis of Multiple Microarray Data Sets Richard Moffitt Georgia Institute of Technology 29 June, 2006 Goal • Use 60 large human microarray datasets. (3924 arrays) • Find reliably coexpressed genes. • http://benzer.ubic.ca/cgi-bin/find-links.cgi – (just google ‘tmm microarray’) Usage Case • Query by gene or probe ID. • Set stringency level. How it Works • Looks for genes that coexpress with the queried-for gene. – correlates gene expression profiles • Stringency requirement eliminates weak links. Our Query • RAP1GSD1, a biomarker form Chang et al • 2 minutes later… Our Results • List of linked genes and some statistics. Visualization • Visualizations of coexpresed gene profiles for each dataset used. Query #2 LETMD1, a biomarker from Citation Spira A, Am J Respir Cell Mol Biol. 2004 Phenotypes_Being_Studied No or mild emphysema, severe emphysema Chip_Platform GPL96: Affymetrix GeneChip Human Genome U133 Array Set HG-U133A for 712X712 Results #2 : Why? • Our first query was from one of the datasets used by Tmm. Synonym Search Conclusion • Useful to make a small list of probable targets. • Useful for some validation? – Similar to GOMiner validation. – Speed will inhibit this. • Semantics is a barrier to usefulness. Acknowledgements • Thanks to: Deepak Sambhara JT Torrance Lauren Smalls-Mantey Malcolm Thomas Randy Han and Kiet Hyun for curating all the biomarker data that was used for test queries.