* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Regulation of Gene Expression
Genomic imprinting wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA interference wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Ridge (biology) wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Messenger RNA wikipedia , lookup
RNA silencing wikipedia , lookup
List of types of proteins wikipedia , lookup
Community fingerprinting wikipedia , lookup
Genome evolution wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transcription factor wikipedia , lookup
Expression vector wikipedia , lookup
Non-coding RNA wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene regulatory network wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene expression wikipedia , lookup
Promoter (genetics) wikipedia , lookup
siswa setyahadi 2014
Regulation of gene
expression
siswa setyahadi 2014
siswa setyahadi 2014
• Gene: A DNA segment that encodes the
all genetic information required to
produce functional biological products
(Note that: not all transcripts are
messenger RNA).
• G.enome: A. complete set of genes of a
given species.
• Gene expression: A process of gene
transcription and/or gene translation
siswa setyahadi 2014
• Most of prokaryotic genomes contain 4.000 or so
genes, although some simple bacteria have only
500-600 genes.
• The human genomes contain about 35.000
genes
Specificity of gene expression
siswa setyahadi 2014
• Temporal specificity (also called stage
specificity)
• Spatial specificity (also called tissue
specificity)
Type of gene expression
a. Constitutive expression
siswa setyahadi 2014
• Some genes are essential and necessary for life, and
therefore are continuously expressed, such as those
enzymes involved in TCA
• These genes are called housekeeping genes.
siswa setyahadi 2014
• b. Induction and repression
• In addition to constitutively expressed genes, all
cells contain genes that are expressed only in
special circumstances
• Such genes are said to be regulated
• Both prokaryotic and eukaryotic cells adapt to
changes in their environment by turning the
expression genes on and off
siswa setyahadi 2014
• Some genes demonstrate higher
expression level once being activated,
such as enzymes involved in DNA
repairing. It is called induced expression .
• On the other hand, some genes are
repressed and their expression levels are
lower, such as the enzymes for Trp
synthesis when Trp is abundant. It is
called repressed expression
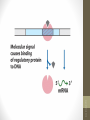
Regulatory Elements
siswa setyahadi 2014
• Gene expression is a multiple-level process.
• Gene expression is also a multiple components
process.
• In general, every step that is required to make an
active gene product can be the focus of a
regulatory event.
• In fact, the most important stage for the regulation
of most genes is when transcription initiation
because it would be a waste to make the RNA if
neither the RNA nor its encoded protein is needed.
Basic Elements
• Basic elements that regulate the transcription
include:
a. Special DNA sequences
b. Regulatory proteins
c. DNA-protein interaction and protein-protein
interaction
siswa setyahadi 2014
d. RNA polymerase
a. Special DNA sequence
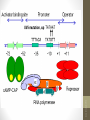
• Gene expression of prokaryotic systems is
regulated based on the operon model which is
composed of structural genes, promoter, operator
and other regulatory sites.
promoter
operator
siswa setyahadi 2014
Other regulatory sites
Structural gene
• Operons
that
include two to six genes transcribed
as a unit are common; some operons contain 20 or more
genes.
• Is a set of adjacent genes whose mRNA is synthesized in
one piece, plus the adjacent regulatory signals that affect
transcription of the genes
Activator
binding site
promoter
A
B
C
Regulatory sequences
Genes transcribed
as a unit
siswa setyahadi 2014
DNA
Repressor
Binding site
(operator)
Types of operons
siswa setyahadi 2014
• Inducible: substrate needs to be present before transcription
of genes involved in its breakdown occurs. Mostly metabolic
pathways-breaking things down for energy, e.g. lac operon
• Repressible: anabolic pathways (building things)- no reason to
make protein to build a molecule that is already available. E.g.
tryptophan operon. An E. coli cell won’t waste a bunch of
energy making tryptophan if it is available from the medium.
Tryptophan represses its own synthesis
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
RNA synthesis is blocked by repressor
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
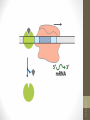
• Promoter. The DNA sequence that RNA-pol can
bind and initiate the transcription.
siswa setyahadi 2014
siswa setyahadi 2014
• As mentioned earlier, RNA polymerases bind to DNA
and initiate transcription at promoters.
• The nucleotide sequences of promoters vary
considerably, affecting the binding affinity of RNA
polymerases and thus the frequency of transcription
initiation.
• By convention, DNA sequences are shown as they
exist in the nontemplate strand, with the 5-terminus
on left
• Most base substitutions in the -10 regions have a
negative effect on function.
• For example, mutations that result in a shift
away from the consensus sequence usually
decrease promoter function; conversely, mutations
toward
consensus
usually enhance promoter
function.
siswa setyahadi 2014
• Most E. coli promoters have a sequence close to a
consensus
• Operator. The DNA sequence adjacent to the
structural genes that the repressor protein can
bind to and prevent the transcription of
structural genes
• The operators are generally near a
promoter.
siswa setyahadi 2014
Coding sequences
b. Regulatory proteins
• For prokaryotic systems:
• Repressor:
• It binds to
the
operator
and prevent the
transcription, known as negative regulation.
• Activator:
siswa setyahadi 2014
• It associates with
DNA near the initiation point,
resulting in the increase of RNA-pol binding affinity and
the enhancement of the transcription efficiency.
siswa setyahadi 2014
• Specific factor: It facilitates the binding of RNA-pol to
particular DNA sequence once associating with other
elements.
• Catabolite gene activator protein (CAP) is a typical
one. For some genes, RNA pol cannot bind to the
promoter without CAP.
siswa setyahadi 2014
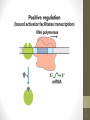
• Regulation by means of a repressor protein that
blocks transcription is referred to as negative
regulation.
• Activators
provide a molecular counterpoint to
repressors; they bind to DNA and enhance
the activity of RNA polymerase at a promoter; this
is positive regulation.
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
• General concepts
• Prokaryotes
• Eukaryotes
• Controls
• Chromosome organization
• Transcription
• Post-transcription
• Translation
• Post translation
siswa setyahadi 2014
oIn prokaryotes, gene expression is controlled by the
external environment. Ex. fuel availability
oIn eukaryotes, gene expression is controlled by the
internal environment. Ex. Hormones, toxic
substances, etc.
siswa setyahadi 2014
Regulation of Gene Expression
in Prokaryotes
siswa setyahadi 2014
• In prokaryotes, control of gene expression occurs at
3 levels:
• Transcription
• Translation
• Post translation
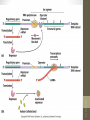
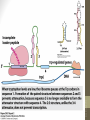
Figure 17-1
Life span (stability)
of mRNA
Translation
rate
Onset of
transcription
Protein activation
or inhibition (by
chemical
modification)
Protein
Ribosome
mRNA
RNA polymerase
RNA polymerase
DNA
Translational control
Post-translational control
siswa setyahadi 2014
Transcriptional control
Common Features
• Prokaryotic systems were developed earlier.
Their internal and external conditions are relative
simple.
• Prokaryotic genes are polycistron systems, that is,
several relevant genes are organized together in a
series format.
siswa setyahadi 2014
• The majority of gene regulation is negative.
• Inducers are used to remove the repression.
siswa setyahadi 2014
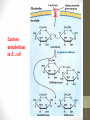
Lac Operon
Lactose metabolism in E. coli
siswa setyahadi 2014
• Galactoside
permease: transport
lactose into the cell
across the cell
membrane
• β -Galactosidase:
hydrolyze lactose to
glucose and
galactose or convert
lactose to allolactose
• Thiogalactoside
transacetylase: may
involved in the
detoxification.
• For lacl· system, all three
enzymes
are
expressed
continuously regardless of
the presence of lactose.
• For lacl+ system,
bacteria do not express
these three enzymes when
glucose is available.
However, bacteria produce
those enzymes if lactose is
present and glucose is
absent
siswa setyahadi 2014
Inducible Expression
siswa setyahadi 2014
• lac gene demonstrate how bacteria use different
types of nutrient as the source of carbon.
• Glucose is the most abundant and prevailing
nutrient, and use of it is the most efficient way.
• Lactose is an alternative choice when glucose is
exhausted.
Sequence of lac operon
• lac operon has a weak promoter
(TTTACA/TATGTT), and has a basal expression
level.
• CAP (Catabolite gene activator protein) binding
site is at - 60 region.
siswa setyahadi 2014
• CAP is a homodimer with binding ability to DNA
and cAMP.
siswa setyahadi 2014
• Glucose inhibits the formation of cAMP.
siswa setyahadi 2014
• When glucose is present, [cAMP] is lower.
• Only after glucose is exhausted, [cAMP] becomes higher.
The CAP-cAMP complex is formed, and this complex binds
to the CAP binding site on lac operon.
siswa setyahadi 2014
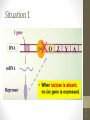
Situation 1
• lac/ gene has its own promoter, and its expression
can produce Lac/ repressor.
• The tetrameric Lac repressor binds to the lac
operator site 01ac·
• The
binding blocks the
RNA-pol moving on
DNA template, and no lacZ, lacY, and lacA are
expressed.
siswa setyahadi 2014
• No use of lactose due to the negative regulation.
siswa setyahadi 2014
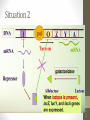
Situation 2
• The galactosidase is weakly
expressed (at the
basal level}.
• When lactose is present, it is converted to
allolactose that binds to the repressor.
siswa setyahadi 2014
• The repressor can no longer bind to the operator,
and lac gene can be expressed.
• Allolactose and IPTG are referred to as inducer.
siswa setyahadi 2014
Inducers
Presence of Lactose
• The lacZ, Y and
siswa setyahadi 2014
A
RNA transcript is very
unstable and could be degraded quickly.
Therefore, the synthesis of three enzymes will
be cease under normal condition.
Situation 3
siswa setyahadi 2014
• When glucose is present, the [cAMP] is low, no
CAP-cAMP is formed and the expression of the
lac operon is still low.
Situation 4
siswa setyahadi 2014
• When glucose is absent and lactose is present,
the CAP-cAMP complex binds to the CAP site to
activate the lac gene.
siswa setyahadi 2014
lac Operon Undergoes Positive
Regulation
Coordinate Regulation
• It is the interaction between CAP-cAMP and RNApol
that greatly stimulates the activation of lac
gene.
• The positive regulation and removal of the
negative regulation of Lac/ repressor allow bacteria
to use lactose as the source of carbon.
siswa setyahadi 2014
• The positive regulation of CAP-cAMP and the
negative regulation of Lac/ repressor constitute a
coordinated regulation unit.
siswa setyahadi 2014
Regulation of Gene Expression
in Eukaryotes
siswa setyahadi 2014
• In eukaryotes, control of gene expression occurs at
these same 3 levels + 2 other levels:
• Chromosome organization (remodeling)
• RNA processing
siswa setyahadi 2014
• Transcription in the eukaryotic nucleus is separated
from translation in the cytoplasm in both space
and time.
• Regulation of eukaryotic gene expression occurs at
multiple levels, including transcription, processing,
mRNA stability, and translation.
• However, like prokaryotic gene, the initiation of
transcription is also a crucial regulation point for
eukaryotic gene expression
Structural Features
siswa setyahadi 2014
• Large genome : 3 x 109 bps, 35,000 genes.
• Monocistron : One gene is transcribed into one
mRNA, and one mRNA then is translated into one
polypeptide.
• Repeated sequences : different lengths and
different frequencies. Often inverted repeats:
complementary and opposite orientation.
• Split genes : separated by introns and exons
alternatively.
Regulation Features
• RNA-pol: 3 forms (I, II, and Ill) for different RNAs
• Changes of chromosomal structure:
• Hypersensitive site
siswa setyahadi 2014
• Base modification : 5°/o of A are methylated.
• Isomers-conversion: from negative supercoil in the
native form to positive supercoil after activation
• Histone changes
• Positive regulation :
• more accurate regulation and more efficient.
• Transcription and translation are separated : at
different locations.
siswa setyahadi 2014
• Post-transcriptional modification : more complicated
than prokaryotes.
• Regulation through intercellular and intracellular
signals :
• hormone.
Gene Regulatory Sequences
• The expression of eukaryotic protein coding
genes is regulated by multiple protein-binding DNA
sequences, generically referred to as
gene
regulatory sequences .
siswa setyahadi 2014
• These include promoters, enhancer and silencer.
Cis-acting Elements
siswa setyahadi 2014
• Note that the gene regulatory sequences are also
called cis-acting elements since they are on the
same DNA molecule as the gene being controlled
(cis is Latin for “on the side”)
• Promoter: TATA box, CAAT box, and GC box
TATA Box
• Sequence: TATAAAA
• Location: - 25-- 30 bp
siswa setyahadi 2014
• Function: It is the binding site for TFII D, which is
required for RNA polymerase binding. Without TATA
box, the 5'-end of transcriptional product is random.
• Sequence: GCCAAT
• Location: - - 80 bp
• Function: It is the binding site for CTF1 (CAATbinding transcription factor) and C/EBP.
The DNA-binding domain of TF1 is rich in basic
Aas, and most likely it is in the alpha-helical
conformation. C/EBP binds to DNA in a dimer
known as leucine zipper.
• Eukaryotes frequently have CAAT boxes, a
strong promoter, usually located around -80
relative to the transcription start site
siswa setyahadi 2014
CAAT Box
GC Box
• Sequence: GGGCGG
siswa setyahadi 2014
• Location: -30 - -110 bp
• Function: It is the binding site for a protein called
Sp1. The DNA-binding domain of Sp1 is near the Cterminus and contains three zinc fingers .
• One or more copies of GC-rich sequences
are also referred to as CpG island.
• These genes, which generally are transcribed
at low rates, have been found upstream from
the transcription start sites of "housekeeping genes."
• Transcription from many eukaryotic promoters can be
stimulated by control elements located thousands of
base pairs away from the start site.
• Such long-distance transcription control elements,
referred to as enhancers, are common in eukaryotic
genomes but fairly rare in bacterial genomes.
• An enhancer is typically 100-200 bp long.
• Enhance has no functionality without promoter.
• Enhances can exert their stimulatory actions over
distances of several thousand base pairs in presence of
promoter.
• They can be upstream, downstream, or even in the
midst of a transcribed gene.
siswa setyahadi 2014
Enhancer
siswa setyahadi 2014
• Their functions are dependent on recognition by
specific transcription factors.
• A specific transcription factor bound at an
enhancer element stimulates transcription by
interacting with RNA polymerase II at a near by
promoter
siswa setyahadi 2014
Silencer
siswa setyahadi 2014
• Silencers are control elements that can inhibit
transcription.
• A specific transcription factor that binds to a silencer
control element and blocks transcription is called a
repressor.
• The protein transcription factors that bind to these
specific sequences are known as trans-acting
factors in that the genes that encode them can be
on different DNA molecules {different
chromosomes).
• Trans-acting proteins bind indirectly to cis-acting
elements and then regulate the transcription
initiation.
• Due to
the
complexity
of
eukaryotic
transcription,
there are many protein factors
in transcription. The transacting factors can be
transcription factors (TF).
siswa setyahadi 2014
Trans-acting Factors
General Structure of TF
• DNA-binding domain
siswa setyahadi 2014
• Activation domain
• Protein-protein interaction domain
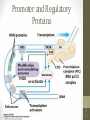
Promoter and Regulatory
Proteins
co-activator
RNA pol II
complex
siswa setyahadi 2014
TFIID
• CTD of RNA-pol II is an important point of interaction
with mediators and other protein complexes.
siswa setyahadi 2014
• Cofactors facilitate the TF assembly.
siswa setyahadi 2014
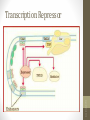
Transcription Repressor
DNA-protein interactions
siswa setyahadi 2014
• Regulatory proteins have discrete DNA-binding
domains of particular structure, i,.e., binding
motif
• They can recognize DNA sequences in a affinity
104-106 times higher than usual
• The AA side chains of regulatory proteins
interact with bases of DNA through H bonds
Motif
• When several
local peptides of defined
secondary structures are close enough in space,
they are able to form a particular "supersecondary " structure.
siswa setyahadi 2014
• Zinc finger
• HLH (helix-loop-helix) HTH (helix-turn-helix)
Leucine zipper
siswa setyahadi 2014
Leucine Zipper
siswa setyahadi 2014
Yeast activator protein in
GCN4 (PDB ID 1YSA)
siswa setyahadi 2014
Zinc Finger
siswa setyahadi 2014
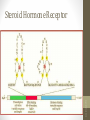
Steroid Hormone Receptor
Gene Specific Regulatory
Proteins
siswa setyahadi 2014
• As mentioned earlier, in order to initiate
transcription, RNA polymerase II requires the
assistance of general (basal) transcription
factors
since no eukaryotic RNA
polymerase
binds
specifically to promoters by itself.
• This binding process involves at least six basal
transcription factors (labeled as TFIIs, transcription
factors for RNA polymerase II).
siswa setyahadi 2014
• Withonly these transcription
(or
basal) factors
and RNA polymerase II attached (the basal
transcription complex), the gene is transcribed at a low or
basal rate.
• However, the rate of transcription can be further
increased by binding of other regulatory DNA binding
proteins to additional gene regulatory sequences (such
as the enhancer regions).
• These regulatory DNA binding proteins are called
gene-specific transcription factors (or transactivators}
because they are specific to the gene involved.
• They also can be called activators, inducers, repressors,
or nuclear receptors.
siswa setyahadi 2014
• In activation of gene, the DNA between the enhancer
and the promoter loops out to allow the
transcription factors bound to the enhancer to interact
with the general transcription factors, other regulatory
proteins or the RNA polymerase itself to increase the rate
of transcription.
• Gene
repressor
proteins
that
inhibit
the
transcription of specific genes in eukaryotes also exist.
For example, when a repressor has attached
to the
silencer region near the enhancers, activators can
be prevented from binding to the enhancers, and
transcription is
• repressed.
• Thus, positive regulations predominate in all systems
characterized of eukaryotes.
siswa setyahadi 2014
• Note that, as already noted, eukaryotic RNA
polymerases
have
little or
no
intrinsic
affinity for their promoters.
• The initiation of transcription is almost always
dependent on the action of multiple activator
proteins.
• One important reason is that the storage of DNA
within
chromatin
effectively
renders most
promoters inaccessible, so genes are normally
silent in the absence of other regulation.
siswa setyahadi 2014
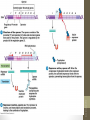
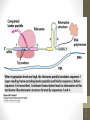
Nucleus
1. Chromatin remodeling
2. Transcription
Chromatin
(DNA-protein complex)
“Open” DNA
(Some DNA not closely
bound to proteins)
Primary transcript
(pre-mRNA)
3. RNA processing
Cap
Tail
Mature mRNA
Cytoplasm
4. mRNA stability
Degraded mRNA
(mRNA lifespan varies)
5. Translation
mRNA
Polypeptide
modification (folding,
transport, activation,
degradation of
protein)
Active protein
siswa setyahadi 2014
6. Post-translational
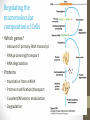
Regulating the
macromolecular
composition of Cells
• Which genes?
• Amount of primary RNA transcript
• RNA processing/transport
• RNA degradation
• Proteins
• translation from mRNA
• Covalent/Allosteric modulation
• Degradation
siswa setyahadi 2014
• Protein modification/transport
1. Principle of gene regulation
Molecular circuits ------------------------------House keeping genes; constitutive gene expression
Inducible; induction; repressible; repression
siswa setyahadi 2014
RNA polymerase binds to DNA at promoters
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
Many prokaryotic genes are clustered and regulated in operons
siswa setyahadi 2014
Lactose
metabolism
in E. coli
siswa setyahadi 2014
The lac operon
The control of gene expression
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
• Each cell in the human contains all the genetic
material for the growth and development of a
human
• Some of these genes will be need to be
expressed all the time
• These are the genes that are involved in of vital
biochemical processes such as respiration
• Other genes are not expressed all the time
• They are switched on an off at need
Operons
• An operon is a group of genes
that are transcribed at the same
time.
• They usually control an important
biochemical process.
• They are only found in
prokaryotes.
Jacob, Monod &
Lwoff
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
© NobelPrize.org
The lac Operon
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
The lac operon consists of three genes each involved in
processing the sugar lactose
One of them is the gene for the enzyme β-galactosidase
This enzyme hydrolyses lactose into glucose and galactose
Adapting to the environment
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
• E. coli can use either glucose, which is a monosaccharide, or
lactose, which is a disaccharide
• However, lactose needs to be hydrolysed (digested) first
• So the bacterium prefers to use glucose when it can
1.
When glucose is present and lactose is absent the E. coli
does not produce β-galactosidase.
2.
When glucose is present and lactose is present the E. coli
does not produce β-galactosidase.
3.
When glucose is absent and lactose is absent the E. coli
does not produce β-galactosidase.
4.
When glucose is absent and lactose is present the E. coli
does produce β-galactosidase
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
Four situations are possible
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
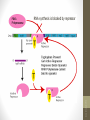
The control of the lac operon
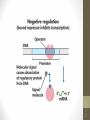
1. When lactose is absent
• A repressor protein is continuously synthesised. It sits on a
sequence of DNA just in front of the lac operon, the Operator
site
• The repressor protein blocks the Promoter site where the
RNA polymerase settles before it starts transcribing
DNA
I
Regulator
gene
© 2007 Paul Billiet ODWS
Blocked
O
Operator
site
z
RNA
polymerase
y
lac operon
a
siswa setyahadi 2014
Repressor
protein
2. When lactose is present
DNA
I
© 2007 Paul Billiet ODWS
O
z
y
a
siswa setyahadi 2014
• A small amount of a sugar allolactose is formed within the
bacterial cell. This fits onto the repressor protein at another
active site (allosteric site)
• This causes the repressor protein to change its shape (a
conformational change). It can no longer sit on the operator
site. RNA polymerase can now reach its promoter site
2. When lactose is present
DNA
I
© 2007 Paul Billiet ODWS
O
z
y
a
Promotor site
siswa setyahadi 2014
• A small amount of a sugar allolactose is formed within the
bacterial cell. This fits onto the repressor protein at another
active site (allosteric site)
• This causes the repressor protein to change its shape (a
conformational change). It can no longer sit on the operator
site. RNA polymerase can now reach its promoter site
3. When both glucose and
lactose are present
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
• This explains how the lac operon is transcribed only when
lactose is present.
• BUT….. this does not explain why the operon is not
transcribed when both glucose and lactose are present.
• When glucose and lactose are present RNA
polymerase can sit on the promoter site but it is
unstable and it keeps falling off
Repressor
protein
removed
DNA
I
O
z
y
a
Promotor site
siswa setyahadi 2014
RNA
polymerase
4. When glucose is absent
and lactose is present
• Another protein is needed, an activator protein. This
stabilises RNA polymerase.
• The activator protein only works when glucose is absent
• In this way E. coli only makes enzymes to metabolise other
sugars in the absence of glucose
Activator protein
steadies the RNA
polymerase
I
© 2007 Paul Billiet ODWS
O
z
y
Promotor
site
a
siswa setyahadi 2014
DNA
Transcription
Summary
Carbohydrates Activator
protein
Repressor
protein
RNA
polymerase
lac Operon
Not bound
to DNA
Lifted off
operator site
Keeps falling
off promoter
site
No
transcription
+ GLUCOSE
- LACTOSE
Not bound
to DNA
Bound to
operator site
Blocked by
the repressor
No
transcription
- GLUCOSE
- LACTOSE
Bound to
DNA
Bound to
operator site
Blocked by
the repressor
No
transcription
- GLUCOSE
+ LACTOSE
Bound to
DNA
Lifted off
Sits on the
operator site promoter site
Transcription
© 2007 Paul Billiet ODWS
siswa setyahadi 2014
+ GLUCOSE
+ LACTOSE
Two Main Mechanisms to Regulate
Transcription in Bacteria
• Use of different factors
• These recognize different classes of promoters
• Allows coordinated expression of different sets of genes
• Binding other proteins (transcription factors) to promoters
These recognize promoters of specific genes
These may bind small signaling molecules
These may undergo post-translational modifications
The protein’s affinity toward DNA is altered by ligand binding or
post-translational modifications
• Allows expression of a specific genes in response to signals in the
environment
siswa setyahadi 2014
•
•
•
•
Regulation by Factors
Sigma 70 consensus– Responsible for the Bulk of mRNA
siswa setyahadi 2014
Sigma 32 consensus– Responsible for “Heat Shock” mRNA
siswa setyahadi 2014
Regulation by Transcription Factors
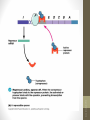
Bacterial Operons
• Operons provide for coordinated expression of genes.
Include promoter, binding sites for activators and
repressors, and functional groupings of genes.
siswa setyahadi 2014
• In this example, A, B, and C are transcribed as one
polycistronic mRNA that is translated into three proteins
•The trp operon
•
Is similar to the lac operon, but functions somewhat
differently
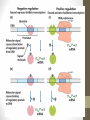
Promoter
Operator
Genes
DNA
Active
repressor
Active
repressor
Tryptophan
Inactive
repressor
Inactive
repressor
Lactose
lac operon
trp operon
siswa setyahadi 2014
Figure 11.1C
Regulation of Gene Expression in Eukaryotes
Not all genes in an organism are “turned on” in all cells or at all
times in particular cell.
All cells of an organism have the same set of genes, but cells
from different tissues look very different and function very
different from one another.
-Development, Stem cells, differentiated cells
siswa setyahadi 2014
- In an adult organism, certain genes are active or inactive
at different times.
siswa setyahadi 2014
Differentiated Cells
Lactose Metabolism
in E. coli
• Lactose is a secondary Carbon
source
• Glucose preferred. Enzymes for
lactose metabolism are
inducible.
• Lactose transport into the cell
siswa setyahadi 2014
• Lactose is hydrolysis into
monosaccharides
The lac Operon has Three Sites
for Binding the Lac Repressor
siswa setyahadi 2014
• Lac Repressor binds with
high affinity to O1(also
O2&3)
• Allolactose reduces
affinity
siswa setyahadi 2014
Model for cooperative binding
• LacR tetramer
siswa setyahadi 2014
• Sugar binding Domain
tetramer “core”
• DNA binding dimer
headpieces
siswa setyahadi 2014
IPTG (lactose analog) causes
subtle conformational change –
loss of affinity
siswa setyahadi 2014
Lac inducer IPTG, structurally similar to lactose
Helix-Turn-Helix Motif is Common in DNABinding Proteins
siswa setyahadi 2014
• One of the helixes (red)
fits into the major groove
of DNA
• Four DNA-binding helixturn-helix motifs (gray) in
the Lac repressor
siswa setyahadi 2014
Binding of Proteins to DNA Often Involves
Hydrogen Bonding
Conformational Change in
Repressor Upon Ligand Binding
• Binding of allolactose or other
lactose analogs, such as IPTG
induces a conformational
change in repressor
• The ligand-bound repressor
dissociates from DNA
siswa setyahadi 2014
• Genes needed for lactose
metabolism are transcribed
Activation of Transcription of
the lac Operon by CRP
• cAMP receptor protein (CRP) is a positive regulator of
the lac operon
• CRP binds to the lac operon in the absence of glucose
siswa setyahadi 2014
• Binding of CRP stimulates expression of the lac operon
CRP-RNAP contacts compensate
for weak RNAP- DNA
siswa setyahadi 2014
• A region of CRP
interacts favorably
with RNA polymerase,
stimulating
transcription of genes
in the lac operon
•
When lactose is low, repressor is bound: inhibition
•
When lactose is high, repressor dissociates permitting transcription
•
When glucose is high, CRP is not bound and transcription is dampened
•
When glucose is low, cAMP is high and CRP is bound: activation
siswa setyahadi 2014
Combined Effects of Glucose
and Lactose on the lac Operon
siswa setyahadi 2014
The trp Operon – Dual Control
Dimeric Trp Repressor Binds to DNA in the
Presence of Tryptophan
siswa setyahadi 2014
• Tryptophan
required for
repression
• Notice that
helix-turn-helix
motifs interact
with DNA via
the major
groove
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
siswa setyahadi 2014
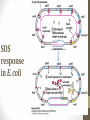
SOS
response
in E. coli
siswa setyahadi 2014
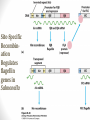
Site-Specific
Recombination
Regulates
flagellin
genes in
Salmonella
siswa setyahadi 2014
Regulation of Transcription in
Eukaryotes
• General transcription factors
• TATA box binding protein (TBP)
• Transcription factors for the assembly of the initiation complex
• Promoter proximal / distal enhancer binding factors
siswa setyahadi 2014
• Homeodomain proteins share similarities with helix-turn-helix
bacterial counterparts nut often involve water bridges between DNA
and protein
• Leucine zippers are made of two amphipathic polypeptides. One side
of each peptide is hydrophobic, facilitating dimerization
• Zinc fingers form elongated loops held together by a single Zn++ ion
Homeodomain Proteins
siswa setyahadi 2014
• Also a helix-turnhelix interacts with
DNA via the major
groove
• One domain of
multidomain –
multiprotein
complex
Leucine Zipper – Dimerization Domains
siswa setyahadi 2014
B (for basic) –Zip proteins
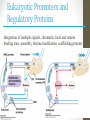
Eukaryotic Promoters and
Regulatory Proteins
siswa setyahadi 2014
Integration of multiple signals, chromatin, local and remote
binding sites, assembly, histone modifcation, scaffolding proteins
siswa setyahadi 2014
Regulation of
transcription of
GAL genes in
yeast
siswa setyahadi 2014
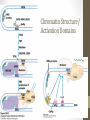
Chromatin Structure/
Activation Domains
siswa setyahadi 2014
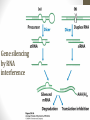
Gene silencing
by RNA
interference
siswa setyahadi 2014
siswa setyahadi 2014
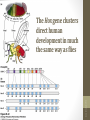
The Hox gene clusters
direct human
development in much
the same way as flies
siswa setyahadi 2014
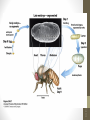
Homeotic mutations transform
one body part into another.
siswa setyahadi 2014
Butterfly colors on a Fruit Fly –
Sean Carrol – U of Wisconsin