* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 2
RNA interference wikipedia , lookup
Protein (nutrient) wikipedia , lookup
List of types of proteins wikipedia , lookup
Epitranscriptome wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene regulatory network wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein adsorption wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Transcriptional regulation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Western blot wikipedia , lookup
DNA vaccination wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Expression vector wikipedia , lookup
Biochemical cascade wikipedia , lookup
Paracrine signalling wikipedia , lookup
Ultrasensitivity wikipedia , lookup
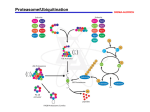
[Vierstra, 2003 TIPS] [Vierstra, 2003 TIPS] Ubiquitin/26S proteasome pathway Simplified Ubiquitination Proteolysis + ATP Ub E1 E2 E3 Target Ub Ub Ub Ub Target + ATP 26S proteasome Loss of 26S proteasome function WT rpn10-1 Smalle et al., 2003 Loss of proteasome function in the rpn10-1 mutant, leads to growth inhibition and the accumulation of polyubiquitinated target proteins. Diversity in Ubiquitination Machinery is largely provided by the many E3s Single E1 Few E2’s Many E3’s E3 structure/function Ub E2 Target protein E3 (Ubiquitin ligase) Target binding E2 binding Ub Target protein E2 Target binding E2 binding E3 (Ubiquitin ligase) E3 structure/function Ub Ub Ub Ub UbUb Ub Ub Target protein 26S proteasome Ub UbUb Ub Target protein E3 Target binding E2 binding Number of E3s per genome Saccharomyces cereviseae 68 Caenorhabditis elegans 657 Drosophila melanogaster 189 Homo sapiens 527 Arabidopsis thaliana 1156 Advantages of proteolysis control in signal transduction 1) Fast response to a change in signal intensity: direct control of protein activities in contrast to transcriptional regulation that involves transcription, transcript processing and translation steps before protein abundance is increased. 2) Proteolysis control can rapidly increase as well as decrease a proteins activity (only an increase is possible with transcriptional regulation). 3) Accurate reflection of signal intensity in response output: secundary modifications such as phosporylation/dephosphorylation can also directly change a proteins activity. However since such controls tend to be leaky, i.e. are the result of modification/demodification equilibria, their outcome depends on the initial abundance of the target protein. Why more E3s in plants? * More E3s means more proteolysis control of signaling. * Energetically wasteful? * Animals can side-step adverse environmental conditions. •The sessile plant must endure. •Plants need to be more sensitive to environmental changes. * Proteolysis control of signaling allows for quick responses to changes in signal intensities (changes in environmental conditions). * Proteolysis control also allows for an accurate responsestrength to signal-intensity ratio. * Allows for a constant state of readiness. * Plants are less energy-limited. From Kepinski and Leyser, 2003 Proteolysis control of signaling Signal transduction leads to destabilization of a repressor of the response or stabilization of a response activator. This is accomplished via secundary modification (phosphorylation or dephosphorylation) of the target protein that leads to or prevents its detection by a Ubiquitin ligase (E3). Alternatively, signaling directly controls E3 affinity for the target protein. Controlling the activity of a protein via its degradation rate allows for faster and more accurate responses to changing concentrations/intensities of the signal (changing environment). The Ub/26SP pathway and signaling Describe two mechanisms that can be used to transform a signal into a response via the regulated degradation of a repressor of this response. Show how increased signal intensity leads to an increased response output. The Ub/26SP pathway and signaling Describe two mechanisms that can be used to transform a signal into a response via the regulated degradation of an activator of this response. Show how increased signal intensity leads to an increased response output. Control of gene expression via conditional proteolysis EXAMPLE 1: Signal (variable) DNA RNA Constitutive expression Response repressor * Response repressor E3 Response (variable) ors e s r n pee se p s ro R Control of gene expression via conditional proteolysis EXAMPLE 2: Signal (variable) DNA RNA Constitutive expression Response activator * Response activator E3 ao r s i t v n pe c e o Rs Response (variable) ABA response (Vierstra, 2009) Control of gene expression via conditional proteolysis EXAMPLE 3: Signal (variable) DNA RNA Constitutive expression Response activator Response (variable) E3 ao r s i t v n pe c e o Rs Control of gene expression via conditional proteolysis EXAMPLE 3: Photomorphogenesis Light (variable) DNA RNA HY5 Constitutive expression COP1 a o i tr sv n pe c e o Rs Light responses (variable) COP1 acts as an E3 to target HY5 for degradation Coil RING E2* WD-40 repeats Ub bZIP COP1 COP1 HY5 HY5 Ub E2 Ub Ub Ub Ub E1 Degradation Degradation via the 26S 26Sby Proteasome proteasome Ub (Osterlund et al.,2000) Photomorphogenesis Light intensity LIGHT COP1 HY5 LIGHT RESPONSES HY5 (Osterlund et al., 2000) Control of gene expression via conditional proteolysis EXAMPLE 4: Signal (variable) DNA RNA Constitutive expression Response repressor Response (variable) E3 ors e s r n pee se p s ro R Control of gene expression via conditional proteolysis EXAMPLE 4: Auxin response pathway Auxin (variable) DNA RNA Constitutive expression AUX/IAA factors TIR1 Auxin Response (variable) ors e s r n pee se p s ro R Auxin response (Vierstra, 2009) Jasmonate response (Vierstra, 2009) Summary: important to remember 1) How does a target protein become polyubiquitinated through the sequential action of E1, E2 and E3 enzymes? 2) 26S Proteasome: structure/function. How does the proteasome detect and then degrade target proteins? 3) Where in the cell does the Ubiquitin/26S Proteasome pathway act? 4) ATP requiring steps in the pathway? Energy is needed to establish specific proteolysis (as opposite to non-specific). 5) Predict the effects of loss of function of different components of the pathway (proteasome --- pleiotropic; E3 --- highly specific phenotype). 6) Why proteolysis control of signal transduction (what are the advantages)? 7) Possible mechanisms of conditional protein degradation to control signal/response ratios (see examples 1-4). 2011 PLS 623 Spring Semester 2011 Exam: Targeted protein Degradation March 25, 2011 Name: _____________________________ E-mail: 1) Describe in detail how a target protein is degraded via the Ubiquitin/26S Proteasome pathway. (20 pts) 2) W hy does protein degradation via the Ubiquitin/26S Proteasome pathway require ATP? (10 pts) 3) W hy are gene families that encode E3s (ubiquitin ligases) more complex (larger) than families that encode E2s or E1s? (10 pts) 4) Describe two mechanisms that can be used to transform a signal into a response via the regulated degradation of a repressor of this response. Show how increased signal intensity leads to an increased response output. (10 pts) 2009 PLS 623 Spring Semester 2009 Exam: Targeted protein Degradation March 27, 2008 Name: _____________________________ E-mail: 1) Describe in detail how a target protein is degraded via the Ubiquitin/26S Proteasome pathway. (20 pts) 2) W hy does protein degradation via the Ubiquitin/26S Proteasome pathway require ATP? (5 pts) 3) I n which cellular compartments is the Ubiquitin/26S Proteasome pathway active? (5 pts) 4) Describe two mechanisms that can be used to transform a signal into a response via the regulated degradation of an activator of this response. Show how increased signal intensity leads to an increased response output. (20 pts) 2008 PLS 623 Spring Semester 2008 Exam: Targeted protein Degradation March 21, 2008 Name_____________________________ 1) Describe in detail how a target protein is degraded via the Ubiquitin/26S Proteasome pathway. (20 pts) 2) W hy are gene families that encode E3s (ubiquitin ligases) more complex (larger) than families that encode E2s or E1s? (10 pts) 3) W hat are the advantages of controlling the abundance of a regulatory protein via targeted proteolysis as compared to transcriptional regulation. (10 pts) 4) Describe two mechanisms that can be used to transform a signal into a response via the regulated degradation of a repressor of this response. Show how increased signal intensity leads to an increased response output. (10 pts)