* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lect21.RegulProtTurnover
Phosphorylation wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Extracellular matrix wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
Endomembrane system wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Signal transduction wikipedia , lookup
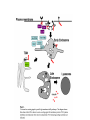
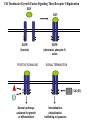
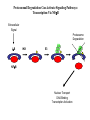
LECT 21: REGULATED PROTEIN TURNOVER Cellular proteins have different stabilities. It is the combination of synthesis and degradation rates that determines the level of a protein in a cell, and changes in either rate can serve as means to regulate a protein’s concentration in the cell. Protein degradation in response to extracellular signals is an important component of some intracellular signaling pathways. Protein degradation is further required to mis-folded or de-folded proteins, which would otherwise be capable of forming insoluble aggregates. Machinery for Protein Degradation Within Cells Cytosolic proteins targeted for degradation are digested in the 26S proteasome, a large multienzymatic structure. Membrane proteins targeted for degradation travel through endosomes to the lysosome, whose lumen contains a collection of proteases. Both targeting processes employ the covalent tagging of proteins with ubiquitin, a small 76 amino acid polypeptide. Cbl Terminates Growth Factor Signaling Thru Receptor Ubiquination EGF EGF EGFR (inactive) POSITIVE SIGNALING EGFR (dimerized, phospho-Y) active SIGNAL TERMINATION Cbl (E3) Several pathways activated for growth or differentiation Internalization, ubiquitination, trafficking to lysosome Proteasomal Degradation Can Activate Signaling Pathways: Transcription Via NFkB Extracellular Signal Proteasome Degradation IkB IKK E3 Ub Ub Ub NFkB Nuclear Transport DNA Binding Transcription Activation Ub Ub Ub