* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 19 - mrswehri.com
RNA silencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Ridge (biology) wikipedia , lookup
Molecular cloning wikipedia , lookup
X-inactivation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Transcription factor wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Non-coding RNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Epitranscriptome wikipedia , lookup
List of types of proteins wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genome evolution wikipedia , lookup
Molecular evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Expression vector wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene regulatory network wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
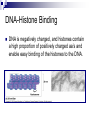
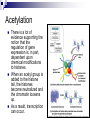
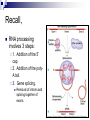
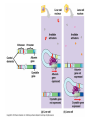
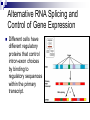
Chapter 19 Eukaryotic Genomes: Organization, Regulation and Evolution. Chromatin The DNA-protein complex found in eukaryotes. It is much more complex in eukaryotes than in prokaryotes. The DNA Within Cells undergoes a variety of changes as it proceeds through the cell cycle. in prophase it’s highly diffuse (thin), but as the cell prepares to divide, it becomes highly condensed. Proteins called histones are responsible for the first level of DNA packing in chromatin. The mass of histone is nearly equal to the mass of DNA. DNA-Histone Binding DNA is negatively charged, and histones contain a high proportion of positively charged aa’s and enable easy binding of the histones to the DNA. DNA-Histone Binding Histones play a very important role in organizing DNA and they are very good at it. Thus, this is a likely reason why histone genes have been conserved throughout the generations in the course of evolution. The structure of histones are very similar among eukaryotes and between eukaryotes and prokaryotes. DNA-Histone Binding and DNA Packing Electron micrographs show unfolded chromatin and they look like beads on a string. These “beads” are referred to as nucleosomes (the basic unit of DNA packing), and the string is DNA. The Nucleosome and DNA Packing A nucleosome is a piece of DNA wound around a protein core. This DNA-histone association remains in tact throughout the cell cycle. Histones only leave the DNA very briefly during DNA replication. With very few exceptions, histones stay with the DNA during transcription. Nucleosome Interaction and DNA Packing The next level of DNA packing takes place between the histone tails of one nucleosome/linker DNA and the nucleosomes to either side. The interactions between these cause the DNA to coil even tighter. As they continue to coil and fold, eventually the DNA resembles that of the metaphase chromosome. DNA Packing Movie http://www.travismulthaupt.com/page1/page5/files/19_02DNAPacking_A.swf Heterochromatin Vs. Euchromatin During interphase, some of the DNA remains condensed as you would normally see it in metaphase. (centrosomes, and some other regions of the chromosome). This is called heterochromatin to distinguish it from euchromatin which condenses and relaxes with the cell cycle. Heterochromatin is rarely transcribed. The Structural Organization of Chromatin The structural organization of chromatin is important in helping regulate gene expression. Also, the location of a gene’s promoter relative to nucleosomes and to sites where DNA attaches to the chromosome scaffold or nuclear lamina can also affect whether it is transcribed or not. Research indicates that chemical modification to the histones and DNA of chromatin influence chromatin structure and gene expression. Acetylation There is a lot of evidence supporting the notion that the regulation of gene expression is, in part, dependent upon chemical modifications to histones. When an acetyl group is added to the histone tail, the histones become neutralized and the chromatin loosens up. As a result, transcription can occur. The enzymes that interact with histones are closely associated with, or are components of transcription factors that bind to promoters. Methylation Addition of a methyl group to a histone tail leads to condensation of the chromatin. Histone Code Hypothesis This hypothesis states that the specific modifications of histones help determine chromatin configuration thus influencing transcription. DNA Methylation DNA methylation is completely separate from histone methylation, but may be a way in which genes become inactivated. Evidence: Inactivated X chromosomes are heavily methylated. In many cells that have inactivated genes, the genes are more heavily methylated than in cells where the genes are active. Control of Eukaryotic Gene Expression Recall the idea of the operon and how it regulated bacterial gene expression. The mechanism of gene expression in eukaryotes is different. It involves chromatin modifications, but they do not involve a change in DNA sequence. Moreover, they can be passed on to future generations by what is known as epigenetic inheritance. Epigenetic Inheritance Epigenetic inheritance occurs when traits are passed on and do not involve the nucleotide sequences (proteins, enzymes, organelles). It also seems to be very important in the regulation of gene expression. The enzymes that modify chromatin are integral parts of the cell’s machinery that regulates transcription. Chromatin Modifying Enzymes These provide initial control of gene expression. They make the region of DNA more or less able to bind DNA machinery. Once optimized for expression, the initiation of transcription is the most universally used stage at which gene expression is regulated. Recall, Eukaryotic genes have promoters, a DNA sequence where RNA polymerase II binds and starts transcription. There are numerous control elements involved in regulating the initiation of transcription. 5’ caps. Poly-A tails. Also, RNA modifications help prevent enzymatic degradation of mRNA, allowing more protein to be made. Movie Recall, RNA processing involves 3 steps: 1. Addition of the 5’ cap. 2. Addition of the polyA tail. 3. Gene splicing. Removal of introns and splicing together of exons. Recall, The transcription initiation complex assembles on the promoter sequence. RNA polymerase II proceeds to transcribe the gene making premRNA. Transcription factors are proteins that assist RNA polymerase II to initiate transcription. RNA Processing Movie Eukaryotic Gene Expression Most eukaryotic genes are associated with multiple control elements which are segments of non-coding DNA that help regulate transcription by binding certain proteins. These control elements are crucial to the regulation of certain genes within different cells. Eukaryotic Gene Expression Only after the complete initiation complex has assembled can the polymerase begin to move along the DNA template strand, producing a complementary strand of DNA. Eukaryotic Gene Expression In eukaryotes, high levels of transcription of a particular gene at the appropriate time depends on the interaction of control elements with other proteins called transcription factors. Enhancers and activators play important roles in gene expression. Enhancers are nucleotide sequences that bind activators and stimulate gene expression. Enhancer-Activator Interaction and Eukaryotic Gene When the activators Expression bind to the enhancers, this causes the DNA to bend allowing interaction of the proteins and the promoter. This helps to position the initiation complex on the promoter so RNA synthesis can occur. Eukaryotic Gene Expression Some specific transcription factors function as repressors to inhibit expression of a particular gene. Certain repressors can block the binding of activators either to their control elements or to parts of their transcriptional machinery. Other repressors bind directly to their own control elements in an enhancer and act to turn off transcription. Transcription Initiation Movie Blocking Transcription Movie Eukaryotic Gene Expression There are only a dozen or so short nucleotide sequences that exist in control elements for different genes. The combinations of these control elements are more important than the presence of single unique control elements in regulating the transcription of a gene. Recall, Prokaryotes typically have coordinately controlled genes clustered in an operon. The operons are regulated by single promoters and get transcribed into a single mRNA molecule. Thus genes are expressed together, and proteins are made concurrently. Control of Eukaryotic Gene Expression Recent studies indicate that within genomes of many eukaryotic species, co-expressed genes are clustered near one another on the same chromosome. However, unlike the genes in the operons of prokaryotes, each of the eukaryotic genes have their own promoter and is individually transcribed. It is thought that the coordinate regulation of genes clustered in eukaryotic cells involves changes in chromatin structure that makes the entire group of genes available or unavailable. Control of Eukaryotic Gene Expression More commonly, co-expressed eukaryotic genes are found scattered over different chromosomes. In these cases, coordinate gene expression is seemingly dependent on the association of specific control elements or combinations of every gene of a dispersed group. Copies of activators that recognize these control elements bind to them, promoting simultaneous transcription of the genes no matter where they are in the genome. Control of Eukaryotic Gene Expression The coordinate control of dispersed genes in a eukaryotic cell often occurs in response to external signals such as hormones. When the steroid enters the cell, it binds to a specific intracellular receptor protein forming a hormone-receptor complex that serves as a transcription activator. Control of Eukaryotic Gene Expression In an alternative mechanism, a signal molecule such as a non-steroid hormone or a growth factor bind to a receptor on a cell’s surface and never enter a cell. Instead, they control gene expression by inducing a signal transduction pathway. Post-transcriptional Regulation and Control of Gene Expression The mechanisms we’ve just discussed involve regulating the expression of the gene. Post-transcriptional regulation involves regulating the transcript after the mRNA has been made. These modes are unique to eukaryotes. Alternative RNA Splicing and Control of Gene Expression Alternative RNA splicing is a way in which different mRNA transcripts are produced from the same primary transcript. This is determined by which RNA segments are treated as introns and which are treated as exons. Alternative RNA Splicing and Control of Gene Expression Different cells have different regulatory proteins that control intron-exon choices by binding to regulatory sequences within the primary transcript. Alternative Mechanisms to Control Gene Expression Protein processing is the final spot for controlling gene expression. Often, eukaryotic polypeptides undergo further processing to yield a functional protein. Regulation can occur at any of the sites of protein modification. Protein Processing Movie http://www.travismulthaupt.com/page1/page5/files/19_10_ProteinProcessing_A.swf