* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Dissection of a DNA-damage-induced transcriptional network using
Transposable element wikipedia , lookup
Oncogenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
X-inactivation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Public health genomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
History of RNA biology wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
Gene nomenclature wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Metagenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene therapy wikipedia , lookup
Primary transcript wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genome editing wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
RNA silencing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
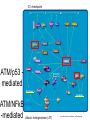
生科4甲 蔡德峰 1 前言 • sequencing of the human -> functional genomics • Gene-expression microarrays and RNA interferences (RNAi) • ATM/NFκB and ATM/p53-mediated arms 2 functional genomics • to gaining system-level understanding of the mechanisms • gene products interact and regulate each other • physiological processes during normal development and in response to homeostatic challenges 3 Gene-expression microarrays 4 • https://www.vbi.vt.edu/ article/articleview/145 RNA interferences (RNAi) (RNA-induced silencing complex) • www.mpg.de/.../ EEB/200432_035.shtml 5 RNA interferences (RNAi) 6 • www.life.uiuc.edu/ shapiro/RNAiApps.html G1 checkpoint ATM/p53 mediated ATM/NFkB -mediated Ataxia- telangiectasia ( AT) 7 • www.biocarta.com/ pathfiles/m_atmPathway.asp NFkB 8 ATM/NFkB -mediated www.mbb.yale.edu/ fl/fl_s_ghosh.htm ATM • NFkB http://www.emdbiosciences.com/popup/cbc/NFKB_Interactive_Pathway.htm ATM/NFkB -mediated 9 Hypothesis • the combined experimental strategy of expression arrays and RNAi is indeed a powerful method for the dissection of complex transcriptional networks, and that computational promoter analysis can provide a strong complementary means for assessing the accuracy of this dissection. 10 實驗流程圖 Microarray analysis 建立siRNA knocked-down cellular systems Computational promoter analysis TRANSFAC GO functional gene annotations Cluster analysis * * * * Database search Definition of the damageresponding gene set * *- New candidate target genes 11 Adapted from Thomas Werner Biomolecular Engineering, 17: 87-94 (2001) 建立siRNA knocked-down cellular systems • Materials and methods DNA fragments To be cloned pSUPER retroviral vector To be transfected HEK293 cell (哺乳動物) (selected with puromycin or hygromycin) 病毒載體用于siRNA表達,其優勢在于可以直接高效率感染細胞進行基因 沉默的研究,避免由于質粒轉染效率低而帶來的種種不便,而且轉染效果更 加穩定。 最適用于:已知一個有效的siRNA序列,需要維持較長時間的基因沉默。 12 以Western blotting 檢驗 RNAi • RNAi并不能完全阻斷基因的表達,特別是表達異常高的基因。 13 Sample preparation and microarray hybridization • Materials and methods HEK293 cell (4 h with 200 ng/ml of NCS.) RNA isolated using TRIzol reagent treated with DNase I phenol/chloroform extracted ethanol-precipitated and quantitated. Affymetrix Human Focus GeneChip arrays 10 種狀態 : five cellular systems (uninfected and the LacZ control cells and cells knocked-down for Rel-A, p53 and ATM), each probed at two time points: without treatment and 4 h after exposure to NCS.14 (All samples were probed in independent triplicates) Computation of gene expression levels from microarray signals • Materials and methods RMA method 1. RMA 計算後, 信號明顯增強 2. RMA 使用齊次多項式證明數據改進更好 15 Definition of the damageresponding gene set • Materials and methods DMA method 取數值at least 1.5-fold in one control (either the uninfected or the LacZ-infected cells), and at least 1.4-fold in the same direction in the other control. A total of 112 genes that were induced in both controls met this criterion and are referred to as the damage-induced gene set. Only seven genes met an analogous criterion for repression in response to NCS treatment 16 Cluster analysis • Materials and methods 112 gene 使用 the EXPANDER package 去 做 average-linkage hierarchical clustering 17 GO functional gene annotations • Materials and methods The gene ontology (GO) annotations Computational promoter analysis • Materials and methods PRIMA software 18 Quantitative real-time RT-PCR • Materials and methods cDNA Five micrograms of total RNA oligo(dT) SuperScript II RNase H- reverse transcriptase real-time PCR 19 20 21 討論 • RNAi and microarray technologies and a recently developed computational tool are powerful • off-target effects • computational promoter analysis was highly enriched for the binding signature of ATF2/ATF3/Jun 22 結論 • RNAi, microarrays and computational promoter analysis 對於 dissection of transcriptional networks 的研究是有力的 • Targeting the primary activator of a DNA damage response network, the upstream regulator(ATM) was indeed required for the induction of much of the network, the two downstream regulators (p53/NFkB)mediated the activation of largely disjoint sets of genes • Statistical tests 聯合 computational promoter analysis 是高精確的 23