* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download (ii) Varshney
Non-coding DNA wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic testing wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic drift wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetically modified crops wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Heritability of IQ wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Human genome wikipedia , lookup
Behavioural genetics wikipedia , lookup
Pathogenomics wikipedia , lookup
Genomic library wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Genetic engineering wikipedia , lookup
Population genetics wikipedia , lookup
Genome editing wikipedia , lookup
Human Genome Project wikipedia , lookup
Minimal genome wikipedia , lookup
Human genetic variation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
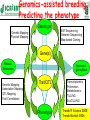
Towards utilization of genome sequence information for pigeonpea improvement By ICAR institutes, SAUs and ICRISAT Pigeonpea (Cajanus cajan L. Millsp) Belongs to family Leguminosae with chromosome no. 2n=22 and genome size of ~833 Mbp A major source of protein to about 20% of the world population (Thu et al., 2003) An abundant source of minerals and vitamins (Saxena et al., 2002) Most versatile food legume with diversified uses such as food, feed, fodder and fuel It is hardy, widely adaptable crop with better tolerance to drought and high temperature Climate change! Pigeonpea – production trends (last five decades) 4.00 3.50 3.00 2.50 Area (M ha) 2.00 Production (M tonnes) 1.50 Productivity (tonnes/ha) 1.00 0.50 0.00 1950-60 1961-70 1971-1980 1981-1990 1991-2000 2001-2007 Years Unfortunately, no increase has been witnessed in its productivity (yield kg ha-1), which in the past five decades has remained stagnant at around 700 kg ha-1 Some constraints in pigeonpea production Sterility mosaic disease (SMD) Fusarium wilt (FW) A route developed and taken by breeders: From germplasm to variety/hybrid Germplasm Superior variety Genomics-assisted breeding: Predicting the phenotype Genotype Genetic Mapping Physical Mapping EST Sequencing Genome Sequencing Map-based Cloning Gene(s) Genetic Resources Genetic Mapping Association Mapping QTL Mapping Trait Correlations Improved germplasm Trait/QTL Transcriptomics Proteomics Metabolomics TILLING EcoTILLING Phenotype Trends Pl Science 2005; Trends Biotech 2006 A variety of approaches (cars) • MAS: MARKER-ASSISTED SELECTION - Plants are selected for one or more (up to 8-10) alleles • MABC: MARKER-ASSISTED BACKCROSSING – One or more (up to 6-8) donor alleles are transferred to an elite line • MARS: MARKER-ASSISTED RECURRENT SELECTION – Selection for several (up to 20-30) mapped QTLs relies on index (genetic) values computed for each individual based on its haplotype at target QTLs • GWS: GENOME-WIDE SELECTION – Selection of genome-wide several loci that confer tolerance/resistance/ superiority to traits of interest using GEBVs based on genome-wide marker profiling Example of development of a submergence tolerant version of Swarna, a widely grown variety, in 2½ years X Swarna: Non-tolerant IR49830-7: Marker-assisted backcrossing tolerant • Target gene selection • Recombinant selection • Background selection Sub1 Swarna-Sub1 Courtesy of David Mackill, IRRI BC2 or BC3 New Sub1 lines (in yellow) and recurrent parents (in white) after 17 days submergence in field at IRRI, 2007DS IR64-Sub1 Samba-Sub1 Samba IR49830 (Sub1) Samba IR64 IR42 IR42 IR64 IR49830 (Sub1) IR49830 (Sub1) IR64-Sub1 IR64 Samba Samba-Sub1 IR64-Sub1 IR42 IR49830 (Sub1) IR42 IR64-Sub1 Samba Samba-Sub1 IR64 Courtesy of David Mackill, IRRI IR49830 (Sub1) Swarna-Sub1 in U.P. (Faizabad area) Courtesy of David Mackill, IRRI, The Philippines Challenges in genomicsassisted crop improvement Narrow genetic base in the primary gene pool Very few molecular (SSR) markers Non-availability of appropriate germplasm such as mapping populations Intraspecific genetic map with low marker density Non-availability of trait-associated markers in breeding Issues of costs and expertise in molecular breeding Germplasm Superior variety Developing infrastructures and sign posts for providing directions (Indo-US AKI, CGIAR-GCP, US-NSF) Gene/transcriptomic/ SNP resources Resource SSRs Pigeonpea 29,000 SNPs GoldenGate 35,000 768 SNPs KASPar assays 1,616 SNPs DArT arrays 15,360 Sanger ESTs 454 /FLX reads TUSs ~20,000 496,705 21,432 Illumina reads (million reads) >160 (14 parents) CMS and mt genome sequencing of pigeonpea Production of A- line seeds Production of hybrid seeds for commercial crop Commercial pigeonpea hybrids production ICPA 2039, ICPB 2039, ICPH 2433 & ICPW 29 sequenced using 454 technology From Orphan crop- genomic resources rich crop Phylogenetic analysis of Cajanus spp. using KASPar assays Cluster-I Cluster-II Cluster-III How to use this genome information… Objectives Molecular mapping of resistance to biotic and abiotic stresses - Mapping populations available and Rf - Genotyping and phenotyping - Marker trait association for resistance to FW, SMD Enhancing the genetic base of pigeonpea genepool by developing multi-parents populations - MAGIC population (2000 lines) developed using 8 parents lines at - NAM population (50 crosses-1000 lines) with 50 parents - High density genotyping or genotyping by sequencing of 3000 - Phenotyping of MAGIC and NAM populations (each population least in 3 environments) Genome wide association studies based on resequencing and phenotyping of germplasm set - Germplasm set of 300-500 lines assembled different - Genotyping-by-sequencing of the germplasm set - Precise phenotyping of the germplasm set by partners - Fine mapping of traits of interest for breeders Bioinformatics analysis to improve the quality of draft genome - Two genome assemblies need to be merged - Defining a consensus genes set - Breeders-friendly genome databases Validation and characterization of 1213 disease resistance genes - Genetic mapping of disease resistance genes - Association of genes with disease resistance traits - Functional validation of selected set of candidate genes resistance - Mining of superior allleles/haplotypes for disease Validation and characterization of ca. 200 abiotic stress tolerance genes - traits Genetic mapping of abiotic stress tolerance genes - Association of genes with abiotic stress tolerance Possible outcomes Superior breeding lines for traits of interest with enhanced genetic diversity Molecular markers associated with resistance to biotic stresses and tolerance to abiotic stresses Alleles and haplotype information available on germplasm set so that breeders can use informative lines Set of well characterized disease resistance and abiotic stress tolerance genes Breeder-friendly genome database of pigeonpea Possible partners NRCPB, New Delhi NBPGR, New Delhi IIPR, Kanpur IARI, New Delhi Uni Agril Sciences- Bangalore Banaras Hindu University ANGRAU- Hyderabad