* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slide 1
Nucleic acid double helix wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
DNA vaccination wikipedia , lookup
Epigenetics wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Designer baby wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Microevolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epitranscriptome wikipedia , lookup
Non-coding DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Transcription factor wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Helitron (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenomics wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
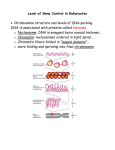
Chapter 32 The Control of Gene Expression in eukaryotes •Background and introduction 1 •Background and introduction Gene regulation in eukaryotes is significantly more complex than in prokaryotes in several ways. 1.The genomes being regulated are significantly larger. The E. coli genome consists of a single,circular chromosome containing 4.6 Mb. This genome encodes approximately 2000 proteins. Saccharomyces cerevisiae (baker’s yeast), 16 chromosomes ranging in size from 0.2 to 2.2 Mb, The yeast genome totals 12 Mb and encodes approximately 6000 proteins. Human cell 23 pairs of chromosomes ranging in size from 50 to 250 Mb. Approximately 23,000 genes are present within the 3000 Mb of human DNA. 2 •Background and introduction 2. Prokaryotic genomic DNA is relatively accessible, eukaryotic DNA is packaged into chromatin, a complex between the DNA and a special set of proteins. The existence of stable cell types is due to differences in the epigenome, differences in chromatin structure, and covalent modifications of the DNA, not in the DNA sequence itself. An electron micrograph of chromatin showing its “beads on a string” character. 3 •Different organs(cells) express different subset of genes 4 •Background and introduction 3. Eukaryotic genes are not generally organized into operons. Instead, genes that encode proteins for steps within a given pathway are often spread widely across the genome. This characteristic requires that other more complex mechanisms function to regulate genes in a coordinated way. 5 Chromatin is made up of nucleosomes which are complexes of DNA and histones Eukaryotic DNA is tightly bound to a group of small basic proteins called Histones which constitute half the mass of a eukaryotic chromosome. The entire complex of a cell’s DNA and associated protein is called chromatin. Histone octamer: The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. Histone octamer An electron micrograph of chromatin 6 showing its “beads on a string” character. DNA wraps around histone octamers to form nucleosomes The four types of histone that make up the protein core are homologous and similar in structure. The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. 7 The eight histones in the core are arranged into a (H3)2(H4)2 tetramer and (H2A–H2B)2 dimers. The DNA forms a left-handed superhelix as it wraps around the outside of the histone octamer(left-handed superhelical ramp). The protein core forms contacts with the inner surface of the DNA superhelix at many points, particularly along the phos-phodiester backbone and the minor groove. Each histone has an amino-terminal tail that extends out from the core structure. These tails are flexible and contain a number of lysine and arginine residues. Covalent modifications of these tails play an essential role in regulating gene expression. Left handed superhelix 8 The DNA double helices are wound around histone core and further packed into higher-order chromatin structure The winding of DNA around the nucleosome core contributes to the packing of DNA by decreasing its linear extent. •An extended 200-bp stretch of DNA would have a length of about 680 Å. Wrapping this DNA around the histone octamer reduces the length to approximately 100 Å along the long dimension of the nucleosome. Thus the DNA is compacted by a factor of 7. •Human chromosomes in metaphase, which are highly condensed, are compacted by a factor of 104. The nucleosomes are arranged in a helical array approximately 360 Å across, forming a series of stacked layers approximately 110 Å apart . The folding of these fibers of nucleosomes into loops further compacts DNA. 9 32.2 Transcription Factors Bind DNA and Regulate Transcription Initiation Eukaryotic transcription factors are different in several ways: 1. DNA-binding sites in prokaryotes are usually quite close to promoters, those in eukaryotes can be farther away from promoters and can exert their action at a distance. 2. Most prokaryotic genes are regulated by single transcription factors, and multiple genes in a pathway are expressed in a coordinated fashion because such genes are often transcribed as part of a polycistronic mRNA. In eukaryotes, the expression of each gene is typically controlled by multiple transcription factors, and the coordinated expression of different genes depends on having similar transcription-factor-binding sites in each given gene. 3. In prokaryotes, transcription factors usually interact directly with RNA polymerase. In eukaryotes, some transcription factors interact directly with RNA polymerase, whereas others interact with other proteins associated with RNA polymerase and still others act by modifying the chromatin structure 10 32.2 Transcription Factors Bind DNA and Regulate Transcription Initiation Eukaryotic transcription factors consist of several domains: The DNA-binding domain binds to regulatory sequences that can either be adjacent to the promoter or at some distance from it. Most commonly, transcription factors include additional domains that help activate transcription. When a transcription factor is bound to the DNA, its activation domain promotes transcription by interacting with RNA polymerase II, by interacting with other associated proteins, or by modifying the local structure of chromatin. 11 A range of DNA-binding structures are employed by eukaryotic DNA-binding proteins Structural motifs for DNA binding domains: 1.Homeodomain: The structure of this domain and its mode of recognition of DNA are very similar to those of the prokaryotic helixturn-helix proteins. In eukaryotes, homeodomain proteins often form heterodimeric structures, sometimes with other homeodomain proteins, that recognize asymmetric DNA sequences A heterodimer motif formed from two different DNAbinding domains, each based on a homeodomain. each homeodomain has a helix-turn-helix motif with one helix inserted into the major groove of DNA 12 Homeodomain structure A range of DNA-binding structures are employed by eukaryotic DNA-binding proteins Structural motifs: 2.Basic-leucine zipper ( bZip) proteins : A pair of long a helices. Part 1: Basic region that lies in the major groove of the DNA and makes contacts responsible for DNA-site recognition. Part2: Coiled-coil structure with its partner. These units are often stabilized by appropriately spaced leucine residues, these structures are often referred to as leucine zippers Coiled-coil structure(part II) Part I This heterodimer comprises two basicleucine zipper proteins(Here only dimer shown). The basic(alkaline ) region lies in the major groove of DNA. 13 Basic-leucine zipper Structural motifs: 3. (Cys2His2) zinc-finger domains : This DNA-binding unit comprises tandem sets of small domains, each of which binds a zinc ion through conserved sets of two cysteine and two histidine residues. DNA-binding domain comprising three zinc-finger domains (shown in yellow, blue, and red) is shown in a complex with DNA. Each zinc-finger domain is stabilized by a bound zinc ion (shown in green) through interactions with two cysteine residues and two histidine residues. Three domains wraps around the DNA in the major groove by domain string. 14 Zinc-finger domains Activation domains (interacting with other proteins) The activation domains generally recruit other proteins that promote transcription. In some cases, these activation domains interact directly with RNA polymerase II or closely associated proteins (indirectly). The activation domains act through intermediary proteins that bridge between the transcription factors and the polymerase. Mediator, a large complex of protein subunits, acts as a bridge between transcription factors activation domains and RNA polymerase II. These interactions help recruit and stabilize RNA polymerase II near specific genes that are then transcribed. DNA binding domain Activation domain 15 Mediator Common features to activation domains 1.They are often redundant; that is, a part of the activation domain can be deleted without loss of function. 2. They are modular and can activate transcription when paired with a variety of DNA-binding domains. 3. Activation domains can act synergistically: two activation domains acting together create a stronger effect than either domain acting separately. Transcrptional repressor like activators, transcriptional repressors act in many cases by altering chromatin structure. 16 Multiple transcription factors interact with eukaryotic regulatory regions Several general transcription factors join with RNA polymerase II to form the basal transcription complex to initiate transcription at a low frequency. For a gene to achieve a higher rate of mRNA synthesis, additional transcription factors must bind to other sites that can be near the promoter or quite distant. This is achieved by combinatorial control. Each factor recruits other proteins to build up large complexes that interact with the transcriptional machinery to activate transcription. Its advantage is that a given regulatory protein can have different effects, depending on what other proteins are present in the same cell. 17 Enhancers can stimulate transcription in specific cell types Transcription factors can often act even if their binding sites lie at a considerable distance from the promoter. These distant regulatory sites in DNA are called enhancers . Enhancers function by serving as binding sites for specific transcription factors. An enhancer is effective only in the specific cell types in which appropriate regulatory proteins are expressed. Protein A Enhancer binding sites Mechanisms: Influence transcription initiation by perturbing the local chromatin structure to expose a gene or its regulatory sites rather than by direct interactions with RNA polymerase Protein A Protein B 18 C Protein Experiment to test enhancers’ fucntion Enhancer controlling the muscle isoform of creatine kinase. This enhancer includes three binding site for regulatory proteins( transcription factors) which are necessary for the enhancer’s function. Protein A Protein A Experiment: Inserting this enhancer near a gene (galactosidase) not normally expressed in muscle cells. Protein B Protein C The result showed that -galactosidase was expressed at high levels in muscle cells of zebrafish embryo but not in other cells. The embyo was treated with X-Gal which was catalyzed by -galactosidase into blue 19 product. -galactosidase in a zebrafish embryo. Application of transcription factors ----Induced pluripotent stem cells (iPS) can be generated by introducing four transcription factors into differentiated cells Pluripotent stem cells have the ability to differentiate into many different cell types on appropriate treatment. Isolated cells derived from embryos show a very high degree of pluripotency. And researchers identified dozens of genes in embryonic stem cells that contributed to this pluripotency. Shinya Yamanaka demonstrated that just four genes out of identified before could induce pluripotency in already-differentiated skin cells(2006 on mouse cells, 2007 on human cells). Finally, Yamanaka introduced genes encoding four transcription factors into skin cells (fibroblasts). Then,the fibroblasts dedifferentiated into cells that appeared to have characteristics very nearly identical with those of embryonic stem cells. That means human can make the already-differentiated cells back to embryonic status with puripotent properties by treatment with related transcription fators. 20 Application of transcription factors ----Induced pluripotent stem cells (iPS) represent powerful new research tools and a new class of therapeutic agents Patient’s fibroblasts could be readily isolated and converted into iPS cells. These iPS cells could then be treated to differentiate into a desired cell type that could then be transplanted into the patient. For example, such an approach might be used to restore a particular class of nerve cells that had been depleted by a neurodegenerative disease. 21 32.3 The Control of Gene Expression Can Require Chromatin Remodeling Chromatin structure plays a major role in controlling eukaryotic gene expression. Relaxing of the chromatin is necessary for gene expression. DNA that is densely packaged into chromatin is less susceptible to cleavage by the nonspecific DNA-cleaving enzyme DNase I. Regions adjacent to genes that are being transcribed are more sensitive to cleavage by DNase I than are other sites in the genome, suggesting that the DNA in these regions is less compacted than it is else-where and more accessible to proteins. We call these sites hypersensitive sites. 22 32.3 The Control of Gene Expression Can Require Chromatin Remodeling Hypersensitive sites are cell-type specific and developmentally regulated. Eg. Hemoglobin genes in the precursors of erythroid cells from 20-hour-old chicken embryos are insensitive to DNase I. However, when hemoglobin synthesis begins at 35 hours, regions adjacent to these genes become highly susceptible to digestion. In tissues such as the brain that produce no hemoglobin, the globin genes remain resistant to DNase I throughout development and into adulthood. It is a necessary for gene expression that chromatin structure change to relax conformation. 23 Experiments clearly revealed the role of chromatin structure in regulating access to DNA binding sites Genes required for galactose utilization in yeast are activated by a transcription factor called GAL4, which recognizes DNA binding sites with two 5’-CGG-3’ sequences on complementary strands separated by 11 base pairs. Approximately 4000 potential GAL4 binding sites of the form 5’-CGG(N)11CCG-3’ are present in the yeast genome, but only 10 of them regulate genes necessary for galactose metabolism. How is GAL4 targeted to only a small fraction of the potential binding sites? GAL4 binding sites (zinc-based domains, 24 not zinc-finger motif) Experiments clearly revealed the role of chromatin structure in regulating access to DNA binding sites Chromatin immunoprecipitation (ChIP) 25 Experiments clearly revealed the role of chromatin structure in regulating access to DNA binding sites ChIP results for analysis GAL4 binding sites Results: Only approximately 10 of the 4000 potential GAL4 sites are occupied by GAL4 when the cells are grow-ing on galactose; More than 99% of the sites appear to be blocked, presumably by the local chromatin structure. Conclusion: not like in prokaryotes all sites appear to be equally accessible, chromatin structure shields a large number of the potential binding sites in eukaryotic cells. Chromatin structure is altered in active genes compared with inactive ones. 26 Steroids and related hydrophobic molecules pass through membranes and bind to DNA-binding receptors Estrogen is one of hydrophobic molecules which can pass through membranes and bind to DNA-binding receptors (Estrogen receptors). Estrogen receptors are members of a large family of proteins that act as receptors for a wide range of hydrophobic molecules, including other steroid hormones, thyroid hormones, and retinoids. The human genome encodes approximately 50 members of this family, often referred to as nuclear hormone receptors. 27 Nuclear hormone receptors have a similar mode of action On binding of the signal molecule (a ligand ), the ligand–receptor complex modifies the expression of specific genes by binding to control elements in the DNA. Estrogen receptors bind to specific DNA sites (referred to as estrogen response elements or EREs) that contain the consensus sequence 5’-AGGTCANNNTGACCT -3’. As expected from the symmetry of this sequence (double chain shows rotatory symmetry), an estrogen receptor binds to such sites as a dimer. 28 Nuclear hormone receptors family has a similar structural features There are two highly conserved domains: a DNA-binding domain and a ligand-binding domain DNA-binding domain: 1. Position: toward the Center 2. Consist of a set of zincbased domains 3. Function: bind to specific DNA sequences by virtue of an a helix that lies in the major groove in the specific DNA complexes formed by estrogen receptors 29 Nuclear hormone receptors family has a similar structural features Ligand-binding domain : 1. Position: C terminal 2. Entirely consist of three layers of helices 3. Function as ligand binding in a hydrophobic pocket that lies in the center of this array of helices 4. Changing conformation when ligand binds to 30 Nuclear hormone receptors family has a similar structural features Ligand-binding domain How does ligand binding lead to changes in gene expression? The binding of ligand alter the DNA-binding properties of the receptor? Like Lac repressor? No 31 Nuclear hormone receptors regulate transcription by recruiting coactivators to the transcription complex Coactivators Because ligand binding does not alter the ability of nuclear hormone receptors to bind DNA, investigators sought to determine whether specific proteins might bind to the nuclear hormone receptors only in the presence of ligand. Such searches led to the identification of several related proteins called coactivators. p160 family(named from their size): SRC-1 ( steroid receptor coactivator-1), GRIP-1 ( glucocorticoid receptor interacting protein-1), and NcoA-1 (n uclear hormone receptor coactivator-1). How to recruit coactivator for nuclear hormone receptor? 32 Coactivators recruitment The binding of ligand to a nuclear hormone receptor induces a conformational change ( helix folds into a groove on the side of the structure on ligand binding) in the ligand-binding domain. This change in conformation generates favorable sites for the binding of a coactivator Coactivator recruitment 33 Coactivators application in drug design ------Steroid-hormone receptors as targets for drugs Molecules such as estradiol that bind to a receptor and trigger signal-ing pathways are called agonists. eg: Athletes sometimes take natural and synthetic agonists of the androgen receptor to enhance the development of lean muscle mass. Other molecules bind to nuclear hormone receptors but do not effectively trigger signaling pathways. Such compounds are called antagonists. eg. Tamoxifen and raloxifene are used in the treatment and prevention of breast cancer, because some breast tumors rely on estrogen-mediated pathways for growth. 34 Coactivators application in drug design ------Mechanism for Tamoxifen anti-breast cacner drugs Tamoxifen binds to the same site as estradiol does. However, tamoxifen has a group that extends out of the normal ligand-binding pocket,as do other antagonists. These groups block the normal conformational changes (helix 12 cannot pack in it usual position to form a groove for coactivator binding) induced by estrogen. So tamoxifen blocks the binding of coactivators and inhibits the activation of gene expression. 35 How do coactivators modulate transcriptional activity? Chromatin structure is modulated through covalent modifications of histone tails Much of the effectiveness of coactivators appears to result from their ability to covalently modify the amino-terminal tails of histones as well as regions on other proteins. eg. some of the p160 coactivators and the proteins that they recruit catalyze the transfer of acetyl groups from acetyl CoA to specific lysine residues in these amino-terminal tails of histone. Histone acetyltransferases (HATs) 36 Histone acetyltransferases (HATs) The histone tails are readily extended; so they can fit into the HAT active site and become acetylated The amino-terminal tail of histone H3 extends into a pocket in which a lysine side chain can accept an acetyl group from acetyl CoA bound in an adjacent site. 37 The consequences of histone acetylation by HATs 1. Lysine bears a positively charged ammonium group at neutral pH. The addition of an acetyl group generates an uncharged amide group. This change dramatically reduces the affinity of the tail for DNA and modestly decreases the affinity of the entire histone complex for DNA, loosening the histone complex from the DNA 2. The acetylated lysine residues interact with a specific acetyllysine-binding domain (bromodomain) that is present in many proteins that regulate eukaryotic transcription 38 Bromodomain and Bromodomain-containing Proteins Bromodomain Bromodomain, comprises approximately 110 amino acids that form a four-helix bundle containing a peptide-binding site at one end. This four-helix-bundle domain binds peptides containing acetyllysine. Notice that an acetylated peptide from histone H4 is bound in the structure. Structure of a bromodomain 39 Bromodomain-containing Proteins Bromodomain-containing proteins are components of two large complexes essential for transcription 1. TAFs(TATA-box-binding protein associated factors) TAF1 contains a pair of bromodomains which are oriented such that each can bind one of two acetyllysine residues at positions 5 and 12 in the histone H4 tail. Thus, acetylation of the histone tails provides a mechanism for recruiting other components of the transcriptional machinery 2. Chromatin-remodeling complexes Utilize the free energy of ATP hydrolysis to shift the positions of nucleosomes along the DNA and to induce other conformational changes in chromatin. Histone acetylation can lead to a reorganization of the chromatin structure, potentially exposing binding sites for other factors. 40 Bromodomain-containing Proteins Chromatin-remodeling complexes Chromatinremodeling complexes Chromatin remodeling 41 Summary for histone acetylation on gene regulation Histone acetylation can activate transcription through a combination of three mechanisms by: 1. Reducing the affinity of the histones for DNA. 2. Recruiting other components of the transcriptional machinery. 3. Initiating the remodeling of the chromatin structure. 42 Acetylation is not the only modification of histones and other proteins in gene-regulation processes. The relation between various histone modifications and their roles in controlling gene expression is sometimes referred to as “the histone code”. K: lysine R: Arginine S:Serine 43 32.4 Eukaryotic Gene Expression Can Be Controlled at Posttranscriptional Levels Just as in prokaryotes, gene expression in eukaryotes can be regulated subsequent to transcription. We shall consider two examples. The first is the regulation of genes taking part in iron metabolism through key features in RNA secondary structure, similar in many ways to prokaryotic posttranscriptional regulation. The second provides an entirely new mechanism. Certain small regulatory RNA molecules allow the regulation of gene expression through interaction with a range of mRNA molecules. 44 Genes associated with iron metabolism are translationally regulated in animals Iron is an essential nutrient ['nju:triənt], required for the synthesis of hemoglobin, cytochromes, and many other proteins. Excess iron can be quite harmful to cells. Animals have evolved sophisticated systems for the accumulation of iron in times of scarcity and for the safe storage of excess iron for later use. RNA secondary structure plays a role in the regulation of iron metabolism in eukaryotes. Key proteins include transferrin, a transport protein that carries iron in the serum, transferrin receptor, a membrane protein that binds iron-loaded transferrin and initiates its entry into cells, and ferritin, an impressively efficient iron-storage protein found primarily in the liver and kidneys. 45 Genes associated with iron metabolism are translationally regulated in animals Ferritin and transferrin-receptor expression levels are reciprocally related in their responses to changes in iron levels. When iron is scarce, the amount of transferrin receptor increases and little or no new ferritin is synthesized. Interestingly, the extent of mRNA synthesis for these proteins does not change correspondingly. Instead, regulation takes place at the level of translation. 46 Ferritin Ferritin mRNA includes a stem-loop structure termed an iron-response element (IRE) in its 5’ untranslated region. This stem-loop binds a 90-kd protein, called an IRE-binding protein (IRP), that blocks the initiation of translation as the IRE binds IRP on the 5’ side of the coding region. When the iron level increases, the IRP binds iron as a 4Fe-4S cluster. The IRP bound to iron cannot bind RNA, because the binding sites for iron and RNA substantially overlap. Thus, in the presence of iron, ferritin mRNA is released from the IRP and translated to produce ferritin, which stores the excess iron. 47 Transferrin Several IRE-like regions are located in 3’ untranslated region on transferrin-receptor mRNA. Under low-iron conditions, IRP binds to these IREs. However, given the location of these binding sites (not 5’), the transferrin-receptor mRNA can still be translated. When the iron level increases and the IRP no longer binds transferrinreceptor mRNA, freed from the IRP, transferrin-receptor mRNA is rapidly degraded. Thus, an increase in the cellular iron level leads to the destruction of transferrin-receptor mRNA and, hence, a reduction in the production of transferrin-receptor protein. 48 Small RNAs regulate the expression of many eukaryotic genes Genetic studies of development in C. elegans revealed that a gene called lin-4 encodes an RNA molecule 61 nucleotides long that can regulate the expression of certain other genes. The 61-nucleotide lin-4 RNA does not encode a protein, but rather is cleaved into a 22nucleotide RNA that possesses the regulatory activity. This discovery was the first view of a large class of regulatory RNA molecules that are now called microRNAs or miRNAs. The key to the activity of microRNAs and their specificity for particular genes is their ability to form Watson–Crick base-pair-stabilized complexes with the mRNAs of those genes. 49 Small RNAs regulate the expression of many eukaryotic genes The miRNAs bind to members of a class of proteins called the Argonaute family to form Argonaute–miRNA complexes which can then bind mRNAs that have sequences complementary to the miRNAs. Once bound, such an mRNA can be cleaved by the Argonaute–miRNA complex through the action of a magnesium-based active site. The miRNAs serve as guide RNAs that determine the specificity of the Argonaute complex 50 MicroRNA–Argonaute complex The miRNA (shown in red) is bound by the Argonaute protein. Notice that the miRNA serves as a guide that binds the RNA substrate (shown in gray) through the formation of a double helix. Two magnesium ions are shown in green. 51 Summary for miRNA in gene regulation This mode of gene regulation is nearly ubiquitous in eukaryotes. Each miRNA can regulate many different genes because many different target sequences are present in each mRNA. 60% of all human genes are regulated by one or more miRNAs The microRNA pathway has tremendous implications for the evolution of gene-regulatory pathways. 52