* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slide 1
Minimal genome wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Primary transcript wikipedia , lookup
Genome evolution wikipedia , lookup
Gene therapy wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
X-inactivation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetic clock wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Oncogenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenomics wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
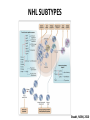
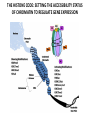
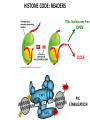
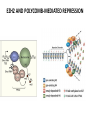
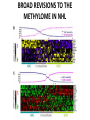
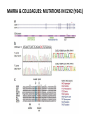
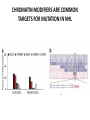
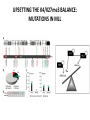
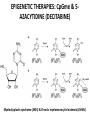
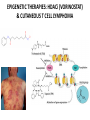
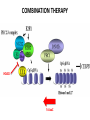
EPIGENETIC MECHANISMS IN B CELL LYMPHOMA Eugene Oltz Dept. of Pathology & Immunology February 21, 2012 LYMPHOMA CLASSIFICATION TYPE INCIDENCE CHARACTERISTICS TREATMENT 5-YEAR SURVIVAL CUTANEOUS T CELL RARE INDOLENT, SMALL LYMPHOID CELLS IN EPIDERMIS STEROIDS, UV, VORINOSTAT 75% MANTLE CELL 3-4% ADULT MALES MODERATELY AGGRESSIVE, LN, SPL, BM INVOLVEMENT, CYCLIN D1 TRANSLOCATION CHEMO (CHOP) 50-70% MALT 5% VARIABLE SIZE & DIFFERENTIATION, VERY INDOLENT EXCISION ~100% HODGKIN 10% REED-STERNBERG CELLS, INFLAMMATION CHEMO (ABVD) / RADIATION >80% DIFFUSE LARGE B CELL 40-50% OLDER ADULTS GERMINAL CENTER CELLS, AGGRESSIVE R-CHOP 60% FOLLICULAR 40% CENTROBLASTS, INDOLENT WATCH & WAIT…THEN RCHOP 75% NHL SUBTYPES Staudt, NEJM, 2010 CNV ANALYSES MOLECULAR DEFECTS IN NHL Staudt, NEJM, 2010 DLBCL SUBTYPES 1. mRNA expression 2. IHC protein expression 3. Copy number aberrations 4. Pathways activated 5. Outcome GCB ABC 1. CD10, BCL6, LMO2+ 2. CD10, BCL2, BCL6 3. generally fewer; t14;18 1. IRF4, BCL2, FOXP1, PIM2 2. CD10, MUM1/IRF4+ 3. generally more; Chr3,18; Bcl2 amplification 4. BCR, NFkB 5. Poor 4. None specific 5. Better Treated with R-CHOP Lenz et al, NEJM 2008 GENE EXPRESSION PROFILES IN NHL SUBTYPES Alizadeh et al., Nature February 2000. GENETICS & EPIGENETICS IN HEALTH & DISEASE GENETICS: SCA CANCER EPIGENETIC: AGING DEVELOPMENT PACKAGING OF GENETIC BLUEPRINTS AS CHROMATIN Closed Heterochromatin Facultative Heterochromatin Open Euchromatin DNA METHYLATION • The 5 position in cytosine can be methylated be DNA methyltransferases (Dnmt) • Some Dnmt’s act during DNA replication to maintain methylation patterns – heritable (others de novo) DNA METHYLATION • When CpG dinucleotides are hypermethylated in a given locus, neighboring genes are usually silent • CpG hypomethylation correlates with gene expression me me HISTONE TAIL MODIFICATIONS • The N-terminal tails of histones protrude out from the nucleosome core • H3 and H4 tails are prime targets for multiple types of covalent modification THE HISTONE CODE: SETTING THE ACCESSIBILITY STATUS OF CHROMATIN TO REGULATE GENE EXPRESSION ChIP and FAIRE-Seq Cross-link whole cells with formaldehyde FAIRE Cells crosslinked with formaldehyde ChIP Sonicate to fragment and immunoprecipitate Perform phenol/ Shear by Sonication Chloroform extraction Histone ChIP Reverse cross-links and purify DNA Library Synthesis End repair, adapter ligation, and amplification Sequencing Illumina massively parallel sequencing Reference chromatin Not crosslinked Perform phenol/ Shear by Sonication Chloroform extraction THE HISTONE CODE • General patterns of histone modifications have been characterized for expressed versus silent genes • Lys-Ac is an active modification • Lys-Me is active (H3K4) or repressive (H3K9 and H3K27), depending on the site • Lys-Ac and –Me are mutually exclusive K4 H4 Ac H3 K4 H4 K27 Repressed Gene ME H3 Ac K27 Active Gene ME EPIGENETICS IN NORMAL DEVELOPMENT • In stem cells, many genes required for differentiation (e.g., Hox) exhibit “bivalent” chromatin that harbors activation AND repressive marks (H3K4me and H3K27me) • Genes with bivalent chromatin are thought to remain in a “poised” state until……. • ……the stem cell receives cues to differentiate down a defined lineage. Chromatin is then modified to a fully active state at lineage-specific genes (H3K4me, H3K9ac) or is fully repressed at genes required for other lineages (H3K9me, H3K27me) THE HISTONE CODE • The epigenetic landscape (pattern of histone modifications) serves as a bar code for many nuclear factors • Three necessary components: writers, erasers, and readers of the code • Covalent modifications of histone tails act as docking sites for reader proteins that: – Stamp new modifications on neighboring nucleosomes – Remodel neighboring nucleosomes – Tether the basal transcription machinery HISTONE CODE: READERS TREs: Nucleosome-Free OPEN CLOSE PIC STABILIZATION HISTONE CODE WRITERS & ERASERS HAT = Histone AcetylTransferase HDAC = Histone DeACetylase • • • • Most histone modifiers are site-specific Numerous HATs and HDACs in mammals Gene regulation HATs: P300 & CBP Some HATs also acetylate non-histone substrates (P53, Rel, Bcl6) HISTONE CODE WRITERS & ERASERS HMT = Histone MethylTransferase HDM = Histone DeMethylase • HMTs and HDMs are usually specific for producing or erasing mono- versus di- versus tri-methylated Lys • H3K4me3 (active promoters): writer =MLL complex; eraser = Jarid 1a, LSD1 • H3K27me3 (repressive): writer = Ezh2 (PRC2); eraser = UTX EZH2 AND POLYCOMB-MEDIATED REPRESSION EPI Hot hypothesis: Changes in epigenome contribute to disease susceptibility, onset, and progression. –Consistent signatures in the epigenetic landscape of diseased cells? –Defects in writers/erasers lead to large-scale revisions to the epigenome and gene expression program DISEASES OF EPIGENETIC ORIGIN: CANCER • Altered epigenomes new gene expression profiles that underlie a broad range of pathologies • The epigenomes of cancer cells are generally CpG hypomethylated (activation of growth genes) but hypermethylated at specific genes (stable repression of tumor suppressors) • Cancer cells overexpress specific subsets of histone code writers (Ezh2 – H3K27me, MLL – H3K4me) BROAD REVISIONS TO THE METHYLOME IN NHL MUTATIONS IN CHROMATIN MODIFIERS ARE PREVALENT IN CANCER MARRA & COLLEAGUES: MUTATIONS IN EZH2 (Y641) MARRA & COLLEAGUES: MUTATIONS IN EZH2 (Y641) MARRA & COLLEAGUES: MUTATIONS IN EZH2 (Y641) MARRA & COLLEAGUES: MUTATIONS IN EZH2 (Y641) POLLOCK & COLLEAGUES Y641 mutations augment K27me2 me3; need WT protein to produce K27me2 NUMEROUS MECHANISMS TO PERTURB K27me3 IN THE EPIGENOME CHROMATIN MODIFIERS ARE COMMON TARGETS FOR MUTATION IN NHL UPSETTING THE K4/K27me3 BALANCE: MUTATIONS IN MLL EPIGENETICS: OPPORTUNITY FOR NEW THERAPEUTIC TARGETS • Unlike genomic lesions, epigenetic changes are reversible • Combination therapies for neurological disorders (bipolar) • A broad range of epigenetic modifiers remain as targets for drug screening 2/9/11 EPIGENETIC THERAPIES: CpGme & 5AZACYTIDINE (DECITABINE) Myelodysplastic syndrome (MDS) & Chronic myelomonocytic leukemia (CMML) EPIGENETIC THERAPIES: HDACi (VORINOSTAT) & CUTANEOUS T CELL LYMPHOMA COMBINATION THERAPY HDACi 5-AzaC COMBINATION THERAPY COMBINATION THERAPY TARGETING THE BCL6/P300 AXIS COMBINATION ONCOGENE/EPIGENETIC THERAPY