* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Objective - Central Magnet School
Genome (book) wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Human genetic variation wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Cancer epigenetics wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Human genome wikipedia , lookup
DNA polymerase wikipedia , lookup
Primary transcript wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genomic library wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA supercoil wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Epigenomics wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Point mutation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Microsatellite wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
SNP genotyping wikipedia , lookup
Helitron (biology) wikipedia , lookup
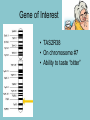
Single Nucleotide Polymorphisms Mrs. Stewart Medical Interventions Central Magnet School Bell Work • Why is Taq polymerase used in PCR instead of human polymerase? • Answer on your own paper Objective • Use laboratory techniques such as DNA extraction, PCR, and restriction analysis to identify single base pair differences in DNA • Explain how single base pair changes called single nucleotide polymorphisms (SNPs) can be identified through genetic testing and often correlate to specific diseases or traits. Single Nucleotide Polymorphisms • The parts of the human genome that vary by just a single nucleotide • Abbreviated as SNPs • Pronounced as “snips” Effect of SNPs • Non-coding DNA regions = no effect • Genes that code for proteins = potentially changing the protein produced E F F E C T –Different phenotypes –Disease/disorder Remember: • a change in DNA can lead to a change in a protein • If the protein plays a role in keeping you healthy, serious consequences may occur. Using a Single Nucleotide Polymorphism to Predict Bitter Tasting Ability Lab Overview Gene of Interest • TAS2R38 • On chromosome #7 • Ability to taste “bitter” What is your phenotype? NonTaster Weak Taster Strong Taster Step 1: Isolate a sample of your DNA from your cheek cells Step 2: Amplifying the Gene of Interest • Using your DNA sample, you will amplify a 220 base pair region of the PTC gene using PCR. – Specific primers attach to either side of the target sequence SNPs for TAS2R38 gene • In this lab, you will investigate one of the base pair changes or single nucleotide polymorphisms (SNPs) that affects a person’s ability to taste the chemical PTC. Genetics Review – Question 1 • The inability to taste PTC is a recessive trait. • If a capital “T” is is used to designate the dominant allele and a lowercase “t” is used to designate the recessive allele, what is the genotype of a “Nontaster”? Answer • A “Nontaster” carries two recessive alleles and thus has the genotype “tt”. Genetics Review – Question 2 • What are the possible genotypes for a “Taster”? Answer • A “Taster” may be homozygous dominant with a genotype of “TT” or heterozygous with a genotype of “Tt”. • In this lab, you will use the tools of molecular biology to determine your genotype for PTC tasting. Step 3: Restriction Analysis • Restriction enzymes, molecular scissors, recognize specific DNA sequences and cut the nucleotide strands. • In this part of the experiment, you will use a specific restriction enzyme, HaeIII, to identify a SNP or base pair difference in the amplified segment of the PTC tasting gene. HaeIII and TAS2R38 gene • HaeIII restriction enzyme – 5’ GGCC – 3’ CCGG 5’ ---GG 3’ ---CC CC--- 3’ GG--- 5’ • TAS2R38 gene variations NONTASTER (tt) TASTER (TT) GGCGGGCACT CCGCCCGTGA GGCGGCCACT CCGCCGGTGA Step 4: Gel Electrophoresis • Gel Electrophoresis separates DNA fragments based on their molecular weight. • Once you have digested your DNA sample with the restriction enzymes, run your product on a gel to analyze your results. What will gel results show? Non-taster Taster























![[edit] Use and importance of SNPs](http://s1.studyres.com/store/data/004266468_1-7f13e1f299772c229e6da154ec2770fe-150x150.png)





