* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Koinophilia wikipedia , lookup
Genome evolution wikipedia , lookup
Population genetics wikipedia , lookup
Genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Oncogenomics wikipedia , lookup
History of RNA biology wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Non-coding RNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genome (book) wikipedia , lookup
Primary transcript wikipedia , lookup
Helitron (biology) wikipedia , lookup
Messenger RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
Microevolution wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Frameshift mutation wikipedia , lookup
Point mutation wikipedia , lookup
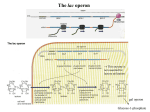
Please feel free to chat amongst yourselves until we begin at the top of the hour. Seminar Agenda Review of Course Information Seminar Discussion Questions Questions & Answers Review of Course Information: Mid-Term Exam This Exam is due by the end of Unit 5 of the course. Worth 100 points. The exam will cover the materials in Units 1-5; it includes everything that was covered in the required readings, lessons, activities, Seminar, and discussion boards. This Exam may only be submitted once and must be taken in one sitting; you may not log-out and log-back in. Exams must be taken during the Unit week (Wednesday to Tuesday). There will be no extensions. Any questions? Discussion Questions 1 If DNA consisted of only two nucleotides (say G and C) in any sequence, what is the minimum number of adjacent nucleotides that would be needed to specify uniquely each of the 20 amino acids? Genetic Code • The genetic code is the list of all codons and the amino acids that they encode • Main features of the genetic code were proved in genetic experiments carried out by F.Crick and collaborators: • Translation starts from a fixed point • There is a single reading frame maintained throughout the process of translation • Each codon consists of three nucleotides • Code is nonoverlapping • Code is degenerate: each amino acid is specified by more than one codon 6 Fig. 8.24 7 Genetic Code • Most of the codons were determined from in vitropolypeptide synthesis • Genetic code is universal = the same triplet codons specify the same amino acids in all species • Mutations occur when changes in codons alter amino acids in proteins 8 Discussion Questions 1 If DNA consisted of only two nucleotides (say G and C) in any sequence, what is the minimum number of adjacent nucleotides that would be needed to specify uniquely each of the 20 amino acids? Discussion Questions 1 If DNA consisted of only two nucleotides (say G and C) in any sequence, what is the minimum number of adjacent nucleotides that would be needed to specify uniquely each of the 20 amino acids? The requirement is that 2n>20, or n=5. If the codons were of four nucleotides, then only 16 amino acids could be specified because 24=16. A codon of five nucleotides does the job because 25=32. In fact, it leaves some extra codons for stop codons and redundancy. Discussion Questions 2 Poly-U RNA codes for polyphenylalanine. If a G is added to the 5’ end of the molecule, the polyphenylalanine has a different amino acid at the amino terminus, and if a G is added to the 3’ end, there is a different amino acid at the carboxyl terminus. What are the amino acids? Discussion Questions 2 Poly-U RNA codes for polyphenylalanine. If a G is added to the 5’ end of the molecule, the polyphenylalanine has a different amino acid at the amino terminus, and if a G is added to the 3’ end, there is a different amino acid at the carboxyl terminus. What are the amino acids? -> NH3-Val-Phe-Phe-Phe-Leu-COOH Codons are read from the 5’ end of the mRNA. The first amino acid in the polypepetide chain is that at amino end, and the last is at the carboxy end. Therefore, a G added to the 5’end of poly-U results in the codon GUU as the first codon in the RNA and hence Valine (val as the first amino acid in the polypeptide. When G is added to the 3’end, it results in a UUG codon, which codes for a leucine (leu) at the carboxyl terminus of the polypeptide. 5’-GUU UUU UUU UUU UUG-3’ Discussion Questions 3 Why are mutations of the lac operator often called a cis-dominant? Why are some constitutive mutations of the lac repressor (lacI) called trans-recessive? Can you think of a way in which a noninducible mutation in the lacI gene might be trans-dominant? Operon Model of Regulation • In bacterial systems, when several enzymes act in sequence in a single metabolic pathway, usually either all or none of the enzymes are produced • This coordinate regulation results from control of the synthesis of one or more mRNA molecules that are polycistronic • Polycistronic mRNAs are transcribed from an operon: a collection of adjacent structural genes regulated by an operator 14 lac Operon • The genetic regulatory mechanism in bacteria was first explained by the operon model of François Jacob and Jacques Monod • They studied lactose-utilization system in E.coli • The lactose-utilization system consists of two kinds of components: structural genes (lacZ and lacY), which encode proteins needed for the transport and metabolism of lactose, and regulatory elements (the repressor gene lacI, the promoter lacP, and the operator lacO) 15 lac Operon • The products of the lacZ (enzyme b-galactosidase) and lacY (transporter lactose permease) genes are coded by a single polycistronic mRNA • The linked structural genes, together with lacP and lacO, constitute the lac operon • The promoter mutations (lacP- ) eliminate the ability to synthesize lac mRNA 16 lac Operon • The product of the lacI gene is a repressor, which binds to a unique sequence of DNA bases constituting the operator • When the repressor is bound to the operator, initiation of transcription of lac mRNA by RNA polymerase is prevented • Because the repressor is necessary to shut off mRNA synthesis, regulation by the repressor is negative regulation 17 lac Operon • Lactose (inducer) stimulate mRNA synthesis by binding to and inactivating the repressor Fig. 9.4a, b 18 lac Operon • In the presence of an inducer, the operator is not bound with the repressor, and the promoter is available for the initiation of mRNA synthesis Fig. 9.4c 19 Discussion Questions 3 Why are mutations of the lac operator often called a cis- dominant? X Mutations of operators are called cis-dominant because, to exhibit their effect, they must be present in the same DNA molecule as the gene they control (that is, the regulatory sequence and the gene must be in a cis configuration). Because the phenotypic effects are observed in partial heterozygotes carrying a normal operator on the other genetic element (for example in an F’lac plasmid), the mutations are said to be cis-dominant.) Discussion Questions 3 Why are some constitutive mutations of the lac repressor (lacI) called trans-recessive? X Constitutive mutations in lacI are called trans- recessive because their effect is overridden by a copy of lacI+ anywhere in the genome (that is they are in a trans configuration relative to the mutant allele). Discussion Questions 3 Can you think of a way in which a noninducible mutation in the lacI gene might be trans-dominant? One can imagine mutations of lacI that bind to the operator irrespective of whether an inducer is present. Such a mutant protein would repress all lac operons, even if normal repressor molecules were present. Such mutations would, therefore, be dominant to the normal repressor. Mutants of this type are actually known: They are call “superrepressor” mutants and are designated lacIs. Any questions?