* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download P. falciparum - University of Notre Dame
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Transposable element wikipedia , lookup
Gene expression programming wikipedia , lookup
Oncogenomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Metagenomics wikipedia , lookup
Genomic library wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genomic imprinting wikipedia , lookup
Designer baby wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Human genome wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Human Genome Project wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genome editing wikipedia , lookup
Pathogenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Chloroplast DNA wikipedia , lookup
Public health genomics wikipedia , lookup
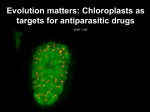
Mother of Green Phylogenomics of the P. falciparum Apicoplast Indiana Center for Insect Genomics An International Center of Excellence University of Notre Dame Purdue University Indiana University Mother of Green • Malaria causes 1.5 - 2.7 million deaths every year • 3,000 children under age five die of malaria every day •Plasmodium falciparum causes human malaria • Drug resistance a world-wide problem • Targeted drug design through phylogenomics P. falciparum Mother of Green • P. falciparum has three genomes Nuclear, mitochondrial, plastid • Animals and insects have only two • Target the third genome • No harm to animals • New antimalarial drug • High risk, high tech, high payoff J. Romero-Severson Department of Biological Sciences Greg Madey Department of Computer Science Mother of Green •Plastids are the third genome •Intracellular organelles •Terrestrial plants, algae, apicomplexans •Functions in plants and algae Photosynthesis Oxidation of water Reduction of NADP Synthesis of ATP Fatty acid biosynthesis Aromatic amino acid biosynthesis •Functions in apicomplexans ? Chloroplast in plant cell plastid Apicoplast in P. falciparum Plastid in Toxoplasma sp. Mother of Green •The apicoplast appears to code for <30 proteins. • Repair, replication and transcription proteins •Why is the apicoplast essential? Mother of Green Phylogenomics • Find the ancestors of the apicoplast • Identify genes in the ancestors • Determine gene function • Look for these genes in the P. falciparum nucleus • Then study regulatory mechanisms in candidate genes Phylogenomics of plastids • Very old lineage (> 2.5 billion years) • Cyanobacterial ancestor • Three main plastid lineages Glaucophytes Group of freshwater algae Chloroplast resembles intact cyanobacteria Chlorophytes Green plant lineage Chloroplast genome reduced Many chloroplast genes now in nuclear genome Rhodophytes Red algal lineage Chloroplast genome bigger than in green plants Oomycetes Apicomplexans One plastid origin Phylogenomics of plastids • One cyanobacterial ancestor ? • Many? • Lineages are not linear Multiple plastid origins Nucleus Primitive eukaryote Endosymbiont plastid Cyanobacteria Nucleus Second eukaryote Nucleomorph Secondary endosymbionts Secondary nonphotosynthetic endosymbiont Plastid disappears The process of endosymbiosis. Horizontal Gene Transfer (arrows) from the plastid to the nucleus. The nucleomorph is a remnant of the original endosymbiont nucleus. Secondary endosymbiont Tertiary endosymbiosis. Horizontal Gene Transfer Third eukaryote Tertiary endosymbionts Plastid disappears Tertiary nonphotosynthetic endosymbiont P. falciparum The information gathering problem • Rapid accumulation of raw sequence information ~100 sequenced chloroplast genomes ~55 sequenced cyanobacterial genomes Rate of accumulation is increasing Information accumulates faster than analyses finish Information in forms not readily accessible • Solution Semi-automated web-services “Smart” web-services The computational problem •Phylogenetic trees NP-hard Poisoned by information conflict •Phylogenies based on individual genes Maximum likelihood models exist Processes are parallelizable Access to compute farms inadequate RAW number-crunching power Greedy • Similar genealogies may be merged Convergence not possible for all Makes computational problem more daunting Candidate genes for deep phylogeny The synthesis of ATP Light-dependent ATP synthesis (photophosphorylation) Hypothesis: Evolution of ATP synthase severely constrained Candidate for ascertainment of deep phylogeny 1st: Individual subunit genealogy 2nd: Merge the data, reanalyze ATP synthase The wheel that powers life Phylogenomics of the P. falciparum Apicoplast • • • • • • • • Extract data from public and private databases Web services Choose a metric for sequence comparison Megablast and others Choose a method to infer genealogy Maximum Likelihood (ML) Develop a strategy to use ML that is feasible fastDNAml and others Create a computational infrastructure Compute farms Dedicated chunks of compute farms Deal with management issues Solve band width problems Convince someone to fund this! Indiana Center for Insect Genomics Mission Create genomics tools for high impact arthropods lacking such tools Develop integrated bioinformatics programs for arthropod genomics Develop specific projects with potential practical application Foster high risk ideas with mini-grants Jeanne Romero-Severson, Director Frank Collins, Co-PI at University of Notre Dame Peter Cherbas, Co-PI at Indiana University Jeff Stuart, Co-PI at Purdue University