* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 投影片 1
Biochemistry wikipedia , lookup
List of types of proteins wikipedia , lookup
Transcription factor wikipedia , lookup
RNA interference wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Community fingerprinting wikipedia , lookup
Molecular cloning wikipedia , lookup
Expanded genetic code wikipedia , lookup
Molecular evolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Genetic code wikipedia , lookup
Point mutation wikipedia , lookup
RNA silencing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Messenger RNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene expression wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Non-coding RNA wikipedia , lookup
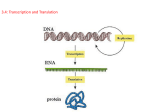
Central Dogma & PCR B91901070 Wang Yu-Hsin Central Dogma What is Central Dogma? DNA RNA Protein transcription translation All cells express their genetic information in this way. Cells must control the amount of protein according to their demand. How do they regulate the efficiency ? Transcription (from DNA to RNA) They use the same language, nucleotide, to store information. Differences between DNA and RNA Deoxyribonucleotide v.s ribonucleotide Base pairs: A = T & C≡G in DNA; T is replaced by U in RNA. Structure in a cell: DNA: Double-stranded helix RNA: Single-stranded with folded shape. Transcription (from DNA to RNA. Cont’d) 1. Open and unwind a small portion of the DNA double-stranded helix. Choose one strand as the template. 2. Produce RNA, with elongating one nucleotide at a time from 5’end to 3’ end and have the sequence exactly complementary to the template. Transcription (from DNA to RNA. Cont’d) There are some differences between RNA transcription and DNA replication. Size of nucleotide sequence. RNA strand does not remain H bond to the DNA template and quickly releases from the DNA as it synthesize for efficient RNA production. RNA polymerase can catalyze the formation of phosphodiester bond( RNA’s sugar-phosphate backbone). But it dose not check the correctness of previous base-pair, which means that RNA transcript can tolerate high error rate. How do they find the site to start or to finish? Transcription (RNA Polymerase) RNA polymerase has to recognize the start of the gene. The initiation of transcription is very important, for it regulates which protein to be produced. Sigma factor in RNA polymerase takes the responsibility of recognizing promoter DNA. After RNA polymerase latches on the DNA and synthesizes about ten nucleotides, sigma factor releases from it. After transcription stops, RNA polymerase leaves and reassociate with a free sigma factor. Then polymerase finds a new promoter… Transcription (in a procaryotic cell) - RNA polymerase first weakly sticks to DNA when it meet the DNA. - Then, polymerase slides along the DNA… - Polymerase finds promoter and latch on the DNA tightly. - Open DNA double-stranded helix and choose one strand as the template. Transcribe RNA by base paring. - When the polymerase encounters the terminal, transcription stop. - Release both the DNA template and newly made RNA. How do eucaryotes work? Transcription (Procaryotes v.s Eucaryotes) Bacteria do not have nucleus, so fresh RNA which are just transcribed can contact rebosomes in the cytoplasm directly. Bacterial mRNA can contain the instructions for several different proteins. Genes are close to each other. Bacterial mRNA degrade rapidly.( about 3 min) Transcription (Procaryotes v.s Eucaryotes. Cont’d) For eucaryotes, newly transcribed RNA( primary RNA), inside the nucleus, needs to take some processes of modification before entering the cytoplasm. Capping at 5’ end with a methyl G. Splicing and remove introns( non-coding sequences). Polyadenylation at 3’end.( poly-A tail) Transcription (Procaryotes v.s Eucaryotes. Cont’d) In introns there are some short nucleotide sequences as cues for its removal. Splicing enzyme: snRNPs, which usually assemble when catalysis begins. With different splicing method, different mRNA forms. This gene recombination provides evolutionary flexibility. Also, the life time of eucaryotic mRNA varies. Translation (from RNA to Protein) They use different languages. mRNA is made up of nucleotides, whereas protein is sequences of amino acid. There are four types of nucleotides; however, there are 20 types of a.a. Three consecutive nucleotides in mRNA as codon. => 64 combinations Genetic codes are general rules for the present-day organism. Translation (tRNA with amino acid) tRNA, transfer RNA About 80 nucleotides long, with 3D structure. Amino acid attaches to 3’ end. Translation (tRNA with amino acid. Cont’d) Redundancy. There are 61 types of codon, 31 types of tRNA, 20 types of amino acid. Different tRNAs can carry the same type of a.a. 2. tRNA can do base-pairing with different mRNA codons. This depends on constructure and means that tRNA can tolerate mismatch (wobble). - Wobbles are alternative codons for a.a, different only in their third nucleotide. 1. Translation (tRNA with amino acid. Cont’d) Aminoacyl-tRNA synthetases: for recognition and attachment of the correct a.a, covalently couple each a.a to its appropriate set on tRNA with high energy bond. Each kind of amino acid has the specific synthetases of its own. To synthesize protein, a cell still needs some place for manufacture... Translation (Ribosomes for protein making) Ribosome: The protein manufacturing machine. Made up of proteins and rRNA. Combination of the large subunit and the small subunit, both made in the nucleus and then sent back to the cytoplasm. Most found in cytosol and ER. Three binding site for tRNA: E, P, and A site. During the process of protein synthesis, no more than two sites contain tRNA mocules at a time. If needed, two subunits come together on mRNA. After protein synthesis complete, they separate. A tRNA carrying the next aa. In the chain bound to A site. Break high energy bond between tRNA and a.a. and form peptide bond in the large subunit. Then... Back to step 1. *What if the initiation site is wrong… Translation (Condons for initiation) Translation (Condons for terminal & others) UAA, UAG, and UGA don’t specify any amino acid. Release factor protein bind to A-site, no more peptidyl transfer but addition of H2O. Free carboxyl end of the growing polypeptide chain. Folding. Polyribosmes. PCR (Polymerase Chain Reaction) - Amplify selected DNA or mRNA sequence of small amount. Entirely in vitro. Based on the use of DNA polymerase to copy a DNA template in repeated rounds of replication. High sensitivity. Polymerase is guide to the sequence to be copied by short primer oligonucleotides. PCR can be used only to clone DNA whose beginning and end are known. (1) 90ºC, open the DNA doublestranded helix. (2) 50ºC, hybridization with primer. (3)75ºC, DNA synthesize. Repeating for 20~30 times. The amount doubles in every cycle. Only the sequence bracketed by two primers is amplified! Reference. Essential Cell Biology, by Alberts et al., 1998, Garland Some pictures are copied from wikipedia and http://www.geneticengineering.org THE END Thanks for your attention