* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download in Power-Point Format
Histone acetylation and deacetylation wikipedia , lookup
Molecular cloning wikipedia , lookup
RNA interference wikipedia , lookup
Gene regulatory network wikipedia , lookup
Messenger RNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Polyadenylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcription factor wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA silencing wikipedia , lookup
Point mutation wikipedia , lookup
Molecular evolution wikipedia , lookup
Epitranscriptome wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Non-coding RNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA polymerase wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
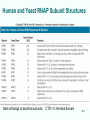
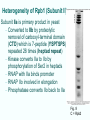
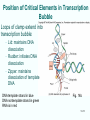
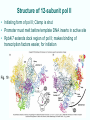
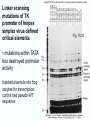
Chapt. 10 Eukaryotic RNA Polymerases and their Promoters Student learning outcomes: • Explain composition of 3 different nuclear RNAPs; emphasis on pol II • Explain which genes each pol transcribes • Describe nature of 3 classes of promoters • Generally appreciate how structural information of pol II has informed mechanistic understanding • Appreciate that mitochondria and chloroplasts also have their own RNA polymerases (not discussed) • Appreciate that Archaeal RNA polymerases share features with 10-1 eukaryotes (not discussed) • Important Figures: 1*, 2, 3, 4, 6*, 9*, 12, 14*, 19*, 20*, 22, 23, 24, 26; Tables: 1, 2, 3 • Review problems: 1, 4, 5, 6, 7, 8, 11, 15, 16, 17, 18, 21, 23, 26, 28; AQ problems: 1, 2, 3, 4 Roger Kornberg: crystal structure of Pol II D4/7 with DNA/RNA active site Nobel Prize 10-2 10.1 Multiple Forms of nuclear RNAP • Nuclei contain 3 RNAP separated by ion-exchange chromatography • Pol I in nucleolus: rRNA genes • Pol II and pol III in nucleoplasm Fig. 1 10-3 Roles of 3 RNA nuclear Polymerases • Pol I large rRNA precursor • Pol II mRNA – Heterogeneous nuclear RNA (hnRNA) – Small nuclear RNA (snRNA) • Pol III tRNAs, 5S rRNA, other small RNA 10-4 Human and Yeast RNAP Subunit Structures Note orthologs to bacterial subunits; CTD = C-terminal domain 10-5 RNA polymerases I, II, III distinguished by sensitivity to inhibitor a-amanitin Fig. 4 pol II is most sensitive Figs. 3 Poisonous Amanita mushroom “death cap” 10-6 Pol II Structure Epitope tagging, purification, helps distinguish true subunits from associated proteins • Add extra domain to one subunit; others normal (e.g., His-tag) • (RNAP labeled by growing cells in labeled amino acids) • Purify complex with antibody to epitope • Denature, separate on SDS-PAG gel • TAP tag common technique (tandem affinity purification) Fig.10-7 6 Pol II Original 10 subunits: • Core – related in structure and function to bacterial core subunits • Common – found in all 3 nuclear RNAP • Nonessential subunits – conditionally dispensable for enzymatic activity • Numbered by size; later subunits 11 and 12 Fig. 7; 32P shows phosphorylated subunits A, Rick Young; b. Roger Kornberg Core Subunits • Rpb1, Rpb2, Rpb3 absolutely required for enzyme activity: deletion mutants are dead; some ts mutants – Homologous to b’-, b-, and a-subunits – Both Rpb1 and b’-subunit bind DNA – Both Rpb2 and b-subunit are at or near the nucleotidejoining active site – Rpb3 does not really resemble a-subunit, but some aa similarity, 2 subunits per holoenzyme Common Subunits Found in all 3 nuclear polymerases: – Rpb5, Rpb6, Rpb8, Rpb10, Rpb12 • All are essential genes (deletion mutants are dead) 10-9 Subunits Nonessential for Elongation • Rpb4 and Rpb7 – – – – Dissociate fairly easily from polymerase substoichiometric quantities Rpb4 may help anchor Rpb7 to pol II Mutants without Rpb4 and Rpb7 transcribe well, but cannot initiate at real promoter • Rpb7 is essential subunit, so must not be completely absent in the rpbD4 mutant • Assist binding of transcription factors 10-10 Review Pol II subunits 10-11 Heterogeneity of Rpb1 (Subunit II) Subunit IIa is primary product in yeast – Converted to IIb by proteolytic removal of carboxyl-terminal domain (CTD) which is 7-peptide (YSPTSPS) repeated 26 times (heptad repeat) – Kinase converts IIa to IIo by phosphorylation of Ser2 in heptads – RNAP with IIa binds promoter – RNAP IIo involved in elongation – Phosphatase converts IIo back to IIa Fig. 8 C = Rpb2 3-D Structure of RNA Pol II - Kornberg Nobel prize • Yeast pol II (pol II D4/7) at 2.8 A: • Deep cleft accepts linear DNA template • Catalytic center at bottom of cleft contains Mg2+ ion • Second Mg2+ ion present in low concentrations • Geometry of linear DNA permits: – TFIID to bind at TATA box of promoter – TFIIB link pol II to TFIID – Places polymerase to – initiate transcription Fig. 10 10-13 3-D Structure - Pol II in Elongation Complex • pol II (D4/7) bound to DNA template, RNA product • Clamp region of pol II closed over DNA and RNA – Closed clamp ensures transcription is processive – transcribe whole gene without falling off, early termination Fig. 13 DNA template strand in blue DNA nontemplate strand in green RNA is in red 10-14 Position of Critical Elements in Transcription Bubble Loops of clamp extend into transcription bubble: – Lid: maintains DNA dissociation – Rudder: initiates DNA dissociation – Zipper: maintains dissociation of template DNA DNA template strand in blue DNA nontemplate strand in green RNA is in red Fig. 14b 10-15 Structure of 12-subunit pol II • Initiating form of pol II; Clamp is shut • Promoter must melt before template DNA inserts in active site • Rpb4/7 extends dock region of pol II; makes binding of transcription factors easier, for initiation Fig. 19 10-16 10.2 Eukaryotic Promoters • Three eukaryotic nuclear RNA polymerases have: – Different structures – Transcribe different classes of genes • Expect the RNAPs recognize different promoters • Emphasis on Pol II (mRNA) 10-17 Class II Promoters Fig. 20 • Promoters recognized by pol II (class II promoters) are similar to prokaryotic promoters: • Considered to have two parts: – Core promoter of 4 elements: TATAAA, TBP, BRE (IIB), – Upstream promoter element TATA box: on nontemplate strand (TATAAA consensus) Very similar to prokaryotic -10 box (TAtAaT consensus)10-18 Linker Scanning mutations clarify promoter elements • Systematically substitute 10-bp linker for 10-bp sequences through promoter • Found mutations within TATA box destroyed promoter activity Fig. 22 10-19 Linker scanning mutations of TK promoter of herpes simplex virus defined critical elements • mutations within TATA box destroyed promoter activity Injected plasmids into frog oocytes for transcription; control had pseudo-WT sequence Fig. 10.23 Core Promoter Elements for pol II • In addition to TATA box, core promoters have: – TFIIB recognition element (BRE) – Initiator (Inr) – Downstream promoter element (DPE) • At least one core element missing in most promoters • TATA-less promoters tend to have DPEs • Promoters for highly specialized genes tend to have TATA boxes • Promoters for housekeeping genes (constitutively active in all cells) tend to lack TATA boxes 10-21 Upstream Elements Regulate pol II • Differ from core promoters in binding to relatively gene-specific transcription factors (Chapt. 12) – GC boxes bind transcription factor Sp1 – CCAAT boxes (‘cat boxes’) bind CTF (CCAAT-binding transcription factor) • Upstream promoter elements can be orientationindependent, yet relatively position-dependent • Linker scanning mutations identified, clarified: – Delete upstream elements -> less expression – Ex. Mutation of GC boxes in SV40 early promoter 10-22 Class I Promoters for rRNA - pol I Fig. 24; linker scanning mutations define sites • Class I promoters are not well conserved sequence • Usually two elements: – Core element surrounding transcription start site – Upstream promoter element (UPE) 100 bp farther – Spacing between elements is important 10-23 Class III Promoters • RNA pol III transcribes a set of short genes • These have promoters that lie wholly within the genes • There are 3 types of these promoters 10-24 Class III Promoters of Pol III Genes are often within the gene • Type I (5S rRNA) has 3 regions: • Type II (tRNA) has 2 regions: • Type III (nonclassical) resemble type II Fig. 26 10-25 10.3 Enhancers and Silencers • Position- and orientation-independent DNA elements stimulate or depress, respectively, transcription of associated genes • Often tissue-specific - rely on tissue-specific DNAbinding proteins for activities • Some DNA elements can act either as enhancer or silencer depending on what is bound to it: – Ex. SV40 Early genes 2 x 72-bp GC boxes Fig. 28 SV40 10-26 Enhancer in immunoglobulin genes Fig. 29 Deletion of enhancer reduces transcription; Fig. 30 enhancer works in either orientation, and position 10-27 Review questions 4. How many subunits does yeast pol II have? Which are core subunits? Which are common to all 3 nuclear RNA polymerases? 7. What is the structure of the CTD of RPB1? 17. Diagram a pol II promoter, showing all of the types of elements it could have. AQ3. You are investigating a new class II promoter. Design an experiment to locate promoter sequences 10-28