* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Arabidopsis thaliana
Genetically modified crops wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Y chromosome wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene desert wikipedia , lookup
Copy-number variation wikipedia , lookup
Genetic engineering wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Non-coding DNA wikipedia , lookup
X-inactivation wikipedia , lookup
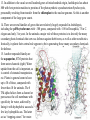
Public health genomics wikipedia , lookup
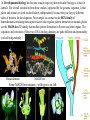
Transposable element wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
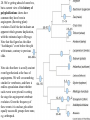
Therapeutic gene modulation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Neocentromere wikipedia , lookup
Genomic imprinting wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genomic library wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Pathogenomics wikipedia , lookup
Human genome wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Helitron (biology) wikipedia , lookup
Human Genome Project wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome editing wikipedia , lookup
IB404 - 13 - Arabidopsis thaliana – Feb 29 1. This small water cress in the mustard family has become the model system for plant biology, with major support from the NSF over the past two decades. Mutant screens became feasible about 15 years ago, and hence much has been learned about development and other aspects of plant biology, eg. circadian rhythms, photo- and geotaxis, basic photosynthetic mechanisms, water regulation, hormonal regulation, defensive secondary metabolism, etc. 2. The genome was sequenced by an international consortium at several labs, and there are too many major players to learn their names. 3. The project was conducted using physically-mapped large BAC and other clones, and the euchromatin was finished in ten large segments for the ten chromosome arms (5 metacentric chromosomes), next slide. 4. The euchromatin arms total around 115 Mbp, while the total genome size is variously estimated as 125-150 Mbp, so there is still a lot of centromeric heterochromatin - hence superficially this genome resembles the Drosophila genome in organization. Indeed among plants it is unusually small. 5. Initial annotation suggested roughly 25,000 genes, although as usual subsequent work using more ESTs, cDNAs, and comparison with the rice genome suggests the number of genes is higher. 6. The basic publications were in 2000, with follow up transcriptome and knockout analyses in 2003, and functional studies on-going. 6. This overview of the genome shows that, like Drosophila, the roughly equal size chromosomes have fairly uniform gene densities on their euchromatic arms (first two lines below chromosome image), but are gene-poor and transposon- and repeat-rich in the centromeric regions (third line). 7. The MT/CP line (fourth) shows the sites of recent insertions of pieces of the mitochondrial and chloroplast genomes, an ongoing process that presumably reflects what has happened continuously since these two endosymbioses occurred ~2 and ~1 BYA. 8. RNAs (last line) are tRNA and snRNAs involved in mRNA splicing. 9. An alternative view of chromosome 2 (right), showing roughly uniform gene content for the chromosome arms, but increasing transposon content (bottom two panels)towards the unsequenced heterochromatic centromere. 10. Sequence organization of the chromosome 5 heterochromatic knob (black blob on the previous slide) (below). Note that the core is a 2.2 kb tandem repeat (yellow tandem arrows). The centromeric heterochromatin is similarly organized, except that the major repeat is 180 bp (size of a nucleosome?). 11. Classification of the proteins into functional categories using the Gene Ontology system shows that about 40% of the functions were unknown, not unusual for such a divergent and new genome. 12. Comparison with other genomes showed how the proportions of the gene complement devoted to certain roles is increased in eukaryotes. Thus the numbers of proteins involved in energy production remain relatively constant (basically these are bacterial after all), while the numbers involved in cell division, transcription, translation, signalling, and intracellular transport are of course much higher in the eukaryotes. 13. In addition to the usual several hundred genes of mitochondrial origin, Arabidopsis has about 800 with best protein matches to proteins of the photosynthetic cyanobacterium Synchocystis, presumably resulting from transfer from the chloroplast to the nuclear genome. So this is another component of the large gene count. 14. There are several families of genes that are relatively hugely expanded in Arabidopsis, including the p450 cytochromes with ~300 genes, compared with ~100 in Drosophila, 75 in C. elegans and only 3 in yeast. In the animals a major role of these proteins is to detoxify the many secondary plant chemicals that serve as defenses against herbivores, as well as other xenobiotics. Ironically, in plants their central role appears to be in generating these many secondary chemicals for defense. 15. Another expanded family are the aquaporins, 8TM proteins that form water channels (right). Water uptake from the soil is important, as is control of stomatal transpiration, etc. Plants in general seem to have up to 30 of these, compared with fewer than 10 for animals. The 8 TM alpha helices form a channel or pore across the cell membrane with specificity for water, achieved by lining it with hydrophobic aas and a few key hydrophilic aas. The latter act as “stepping-stones” for water. 16. Developmental biology has become a major topic in plant molecular biology as it has for animals. The overall conclusion from these studies, supported by the genome sequence, is that plants and animals evolved multicellularity independently because they use largely different suites of proteins for development. For example, in contrast to the HOX family of homeodomain-containing transcription factors that regulate pattern formation in animals, plants use the MADS-box TF family that mediate pattern formation in flowers and other organs. The sequences and structures of these two DNA-binding domains are quite different and presumably evolved independently. Homeodomain MADS box Some MADS box mutants - wild type is on left. 17. A major feature of the genome is the presence of large apparently duplicated regions, both between and within chromosomes. These are thought to be the vestiges of a polyploidization event roughly 30 Myr ago. Further analysis actually suggests another even older polyploidization event, so the combination of these events, and differential retention of duplicated genes, explains much of the large gene count of >25,000. 18. Polyploidization events can be detected in several ways. A second way is to plot a histogram of the divergences of pairs of paralogous genes within a genome, usually using the silent substitution frequency or Ks. As shown below, this should reveal a peak interrupting an otherwise steady decay of gene pairs and sequence identity with time. 19. Polyploidization events can be dated by comparing histograms of Ks for pairs of genes within a species (paralogs) to pairs of genes (orthologs) between species. Splitting the Arabidopsis gene pairs into recent blocks of duplicates and old blocks of duplicates allows the timing to be estimated by comparisons with the Ks values with other plants, e.g. young Brassica, older Medicago and tomato, and much older rice (a monocot in comparison with these dicots) (right). The vertical dotted lines are the peaks of Ks distributions for the younger and older polyploidizations in Arabidopsis. 20. We’re getting ahead of ourselves, but a current view of the history of polyploidizations shows how common they have been in angiosperm (flowering plant) evolution. Each blue dot indicates an apparent whole genome duplication, with the estimated age in Myr ago. Note that this figure has the older “Arabidopsis” event before the split with tomato, contrary to previous slide. Note also that there is a really ancient event hypothesized at the base of angiosperms. We will see something similar for vertebrates, and there is endless speculation about whether such events were pivotal in setting the stage for angiosperm/vertebrate evolution. Given the frequency of these events it is unclear, plus other equally successful groups show none, e.g. arthropods.