* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Introduction to GO Annotation
Epigenetics of neurodegenerative diseases wikipedia , lookup
Protein moonlighting wikipedia , lookup
Metagenomics wikipedia , lookup
Point mutation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Public health genomics wikipedia , lookup
Copy-number variation wikipedia , lookup
Pathogenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
The Selfish Gene wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome editing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene nomenclature wikipedia , lookup
Microevolution wikipedia , lookup
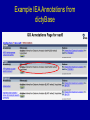
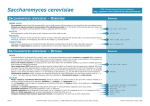
GO Annotations: What are they and how are they made? Rama Balakrishnan Saccharomyces Genome Database Stanford University Why GO? • Comparisons between species – Gets powerful if we have broader species coverage • Terminology used to describe your species becomes more accessible • Genome-wide analyses – Can be used to figure out if there is anything common in your microarray cluster How can this be accomplished? • By providing Annotations (i.e. link genes to GO terms) • By providing Content (controlled vocabularies to the ontologies) • Sharing tools, scripts and other resources I want to Annotate my Genome. Where do I start? • GO website http://www.geneontology.org – Check the Annotation Documentation, Teaching Resources section on the GO website • http://www.geneontology.org/GO.current.annotations.shtml • http://www.geneontology.org/GO.teaching.resources.shtml – – – – Attend one of the annotation camps Annotation mailing list Source Forge tracker for annotation related issues Farmanimals mailing list (new) • A GO consortium member can mentor a new comer if need be What tools/infrastructure do you need to record annotations? • Excel spread sheet (simple, easy, small scale) OR • FileMaker Pro, Access – Simple databases, scales very well • ORACLE or MySQL Lets Get Started! • What is an annotation? • Annotation approaches • Strategies for identifying literature to annotate • Strategies for reading a paper for annotation • Strategies for annotating a gene and a genome What is a GO annotation? • A annotation is a piece of information associated with a gene product • A gene product is usually a protein but can be a functional RNA • A GO annotation is a Gene Ontology term associated with a gene product Approaches for annotation of a genome 1. Automated/Electronic approaches 2. Manual approaches 3. Combinatorial approach Anatomy of a GO annotation Reference Gene Product IMP, IGI, IPI, ISS, IDA, IEP, TAS, NAS, ND, RCA, IC, IEA Evidence Code GO Term Literature Source 1. PubMed - National Library of Medicine, National Institutes of Health - http://ncbi.nlm.nih.gov 2. Agricola - United States Department of Agriculture, National Agricultural Library - http://agricola.nal.usda.gov 3. Embase - Elsevier - http://www.embase.com 4. Biosis - Thomson - http://www.biosis.org 5. Unpublished - abstract in your own database - unpublished abstract submitted to GO references collection Evidence types • • • • • • • • • • ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available • IEA: Inferred from electronic annotation Electronic Annotation • First pass annotations relatively quickly • Annotation derived without human validation – Sequence similarity, e.g. Blast search ‘hits’ – Mapping file, e.g. interpro2go, ec2go, etc. • Useful For: – genomes that don’t have extensive literature – groups with limited curatorial resources Electronic Annotation • Typically based on sequence similarity • Document the method used in a abstract • Internal Reference - unpublished abstract in your own database - unpublished abstract submitted to GO references collection • Annotation is not reviewed by human • IEA evidence code Example IEA Annotations from dictyBase Example unpublished reference Manual annotation • • • • Created by scientific curators Time intensive Utilizes published literature Manatee (offered by TIGR) Combinatorial Approach, e.g. using sequence similarity 1. Alignments published in literature 2. Analysis using full length protein 3. Analysis using protein domains Additional annotation information • WITH/FROM: describes the evidence code – IPI, IGI, IMP, IEP, ISS, IEA, IC, NAS – Contains the interacting or similar gene product • QUALIFIER: describes the GO term – NOT – contributes to – colocalizes with Example Annotation nek2 PMID: 11956323 Reference Gene Product IDA centrosome GO:0005813 Inferred from Direct Assay GO Term Evidence Code What to Search For in Published Literature? 1. Species name 2. Gene/gene product names: daf-12, spo11, Sonic hedgehog 3. Process AND species: embryonic development AND elegans 4. Function AND species: transcription factor AND mays 5. Cellular component AND species (genus): plasma membrane AND Drosophila GO Annotation: GMOD Tools for Enhancing Information Retrieval GMOD – Generic Software Components for Model Organism Databases - http://www.gmod.org/home - Literature search tools: PubSearch – http://www.gmod.org/?q=node/44 PubFetch - http://www.gmod.org/?q=node/84 Textpresso – http://www.textpresso.org - full text of articles - semantic categories GO Annotation: Strategies for Identifying Literature for Curation 1. Primary research literature with new experimental data - Mutant phenotypes – process - Activity assays – function - Localization studies – component 2. Computational analyses - Phylogenetic analysis – function (ISS) - Domain analysis 3. Review articles - TAS evidence Which parts of the paper are most important? • Introductory information • Abstract • Experimental Results • Results: Figures, Tables, Text • Materials and Methods • Explanatory text (use with caution) • (Introduction) – mostly TAS information • (Discussion) How is it different from reading papers as a bench scientist? • Don’t be swayed by the speculations or theories that may appear in the Discussion. • Focus on the actual results vs. the possible implications of those results. • Read for details and contact authors if key identifiers are missing. How to search for a GO term? • Web based tools– AmiGO browser (http://www.godatabase.org) – QuickGO (http://www.ebi.ac.uk/ego/) • Downloadable tool (https://sourceforge.net/projects/geneontology/) – OBO-Edit – Download the ontology file Extracting Information from a paper Sample text from PMID: 12374299 In this study, we report the isolation and molecular characterization of the B. napus PERK1 cDNA, that is predicted to encode a novel receptor-like kinase. We have shown that like other plant RLKs, the kinase domain of PERK1 has serine/threonine kinase activity, In addition, the location of a PERK1-GTP fusion protein to the plasma membrane supports the prediction that PERK1 is an integral membrane protein…these kinases have been implicated in early stages of wound response… Example Manual Annotations from SGD Strategies for annotating • Approaches • Updating GO annotations Annotation from published literature 1. Focus on known genes 2. Identify literature relevant to that gene a. using gene names, species name 3. Complete annotation set for a gene a. annotate available experimental data b. annotations to root nodes indicate nothing is known Updating GO annotations • Ongoing process – New experimental data – More specific annotation • Replace obsolete terms • Rerun computational methods – InterProScan and interpro2go I don’t see terms in the ontology that describe the biology of my species. • Send an email to the GO mailing list • Source Forge (SF) tracker for term related issues https://sourceforge.net/projects/geneontology/ • Content meetings – Organized by the consortium if the ontology related issues can’t be resolved over email/SF – Look for announcements on the GO website, mailing lists I have my annotations, what next? • • • Prepare to submit your annotations to the GO consortium Follow file format Information on Annotation file format can be found at: http://www.geneontology.org/GO.annotation.shtml#file • A file in this format is called gene_association file DB: Source of the ID in column 2 Examples- SGD, MGI, UniProt ID for the gene or gene_product Examples - FBgn0015331, MGI:99240, SPAC9.03c Symbol like Brr2, DDX21_HUMAN that means something to a biologist, not an ID Object_Type - gene, transcript, protein, protein_structure, or complex, should match the ID Sample gene-associations file Optional column DB source SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD DB Object ID S000004660 S000004660 S000004660 S000004660 S000000289 S000000289 S000000289 S000000289 S000000289 S000003916 S000003916 S000003916 S000005275 S000005275 S000005275 S000005525 S000005525 S000005525 S000001837 S000001837 S000001837 S000000704 S000000704 S000000704 Object Symbol Qualifier GOID AAC1 GO:0005743 AAC1 GO:0005471 AAC1 GO:0006839 AAC1 GO:0009060 AAC3 GO:0005743 AAC3 GO:0005471 AAC3 GO:0009061 AAC3 GO:0009061 AAC3 GO:0009061 AAD10 GO:0008372 AAD10 GO:0018456 AAD10 GO:0006081 AAD14 GO:0008372 AAD14 GO:0018456 AAD14 GO:0006081 AAD15 GO:0008372 AAD15 GO:0018456 AAD15 GO:0006081 AAD16 GO:0008372 AAD16 GO:0018456 AAD16 GO:0006081 AAD3 GO:0008372 AAD3 GO:0018456 AAD3 GO:0006081 DB:reference SGD_REF:S000050955|PMID:2167309 SGD_REF:S000050955|PMID:2167309 SGD_REF:S000050955|PMID:2167309 SGD_REF:S000050955|PMID:2167309 SGD_REF:S000045889|PMID:2165073 SGD_REF:S000045889|PMID:2165073 SGD_REF:S000045889|PMID:2165073 SGD_REF:S000052497|PMID:1915842 SGD_REF:S000045889|PMID:2165073 SGD_REF:S000069584 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000069584 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000069584 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000069584 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000069584 SGD_REF:S000042151|PMID:10572264 SGD_REF:S000042151|PMID:10572264 Optional column Ev_code TAS IDA IGI IGI ISS ISS IGI IGI IEP ND ISS ISS ND ISS ISS ND ISS ISS ND ISS ISS ND ISS ISS With/From Aspect DB object Name Synonym C ADP/ATP translocator YMR056C F ADP/ATP translocator YMR056C SGD:S000000126 P ADP/ATP translocator YMR056C SGD:S000000126 P ADP/ATP translocator YMR056C SGD:S000000126|SGD:S000004660 C ADP/ATP translocator YBR085W|ANC3 SGD:S000000126|SGD:S000004660 F ADP/ATP translocator YBR085W|ANC3 SGD:S000000126 P ADP/ATP translocator YBR085W|ANC3 SGD:S000000126|SGD:S000004660 P ADP/ATP translocator YBR085W|ANC3 P ADP/ATP translocator YBR085W|ANC3 C aryl-alcohol dehydrogenase YJR155W(putative) F aryl-alcohol dehydrogenase YJR155W(putative) P aryl-alcohol dehydrogenase YJR155W(putative) C aryl-alcohol dehydrogenase YNL331C(putative) F aryl-alcohol dehydrogenase YNL331C(putative) P aryl-alcohol dehydrogenase YNL331C(putative) C aryl-alcohol dehydrogenase YOL165C(putative) F aryl-alcohol dehydrogenase YOL165C(putative) P aryl-alcohol dehydrogenase YOL165C(putative) C YFL057C F YFL057C P YFL057C C aryl-alcohol dehydrogenase YCR107W (putative) F aryl-alcohol dehydrogenase YCR107W (putative) P aryl-alcohol dehydrogenase YCR107W (putative) Object_type gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene gene Taxon ID taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 taxon:4932 Date 20010118 20010213 20040226 20040226 20040226 20040226 20040226 20040226 20040226 20010119 20020902 20020902 20010119 20020902 20020902 20010119 20020902 20020902 20020902 20020902 20020902 20010119 20020902 20020902 Assigned by SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD SGD How do I share my gene_associations file? • Provide them to the larger community by submitting your annotations to the GO project • What information should I submit to GO? – gene-association file – Contact email address • Where should I submit the data? – Send the file to Mike Cherry or send an email to the GO mailing list – [email protected] Databases contributing annotations – dictyBase (Dictyostelium discoideum) – FlyBase (Drosophila melanogaster) – GeneDB (Schizosaccharomyces pombe, Plasmodium falciparum, Leishmania major and Trypanosoma brucei) – UniProt Knowledgebase (Swiss-Prot/TrEMBL/PIR-PSD) and InterPro databases – Gramene (grains, including rice, Oryza) – Mouse Genome Database (MGD) and Gene Expression Database (GXD) (Mus musculus) – Rat Genome Database (RGD) (Rattus norvegicus) – Reactome – Saccharomyces Genome Database (SGD) (Saccharomyces cerevisiae) – The Arabidopsis Information Resource (TAIR) (Arabidopsis thaliana) – The Institute for Genomic Research (TIGR): databases on several bacterial species – WormBase (Caenorhabditis elegans) – Zebrafish Information Network (ZFIN): (Danio rerio) Species coverage • All major eukaryotic model organism species • Human via GOA group at UniProt • Several bacterial and parasite species through TIGR and GeneDB at Sanger – many more in pipeline Annotation coverage Annotation coverage Resources the GO project offer to help you get started • GO website – http://www.geneontology.org – Lots of documentation – Tools, tutorials and software • GO mailing list • [email protected] • GO project on Source Forge (SF) – https://sourceforge.net/projects/geneontology/ • AmiGO web application (http://www.godatabase.org) • GO database