* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Imprinted Genes
Public health genomics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA vaccination wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of depression wikipedia , lookup
X-inactivation wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetic clock wikipedia , lookup
DNA methylation wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Oncogenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Epigenomics wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Epigenetics wikipedia , lookup
Designer baby wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
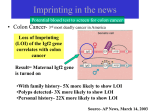
Epigenetic Programming in utero and Later in Life Disease CNRU Retreat September 28, 2006 Ken Eilertsen, Ph. D. Stem Cell Biology Group/Epigenetics and Nuclear Reprogramming Outline • Brief overview of where we started and how we got here – How we got here is really a function of the CNRU enabling us to ask new questions. • A summary of our first experiment venturing into this area • Overview of the current P & F award Past 10 years… • My lab focused on assisted reproductive technologies research • Primarily somatic cell nuclear transfer (cloning) – Nuclear reprogramming • Upon arriving at The Pennington, we wanted to explore new, but very related questions. – Barker Hypothesis – CNRU has been key to meeting this latter objective. Barker Hypothesis Fetal (Developmental) Origins of Adult Disease Hypothesis: •Posits that a poor in utero environment elicited by maternal dietary or placental insufficiency ‘programs’ susceptibility in the fetus to later development of cardiovascular and metabolic disease. Programming: -commonly ascribed to any situation where a stimulus or insult during development establishes a permanent physiological response. -in the context of a predisposition to a later in life disease, no mechanism described. Some animal models lend support to Developmental Origins of Disease Hypothesis A. B. Maternal low protein diet causes a significant reduction in ICM (embryonic stem cells) and TE (placenta) cell numbers at blastocyst stage. Assisted Reproductive Technologies (ART) in humans have been linked to later in life disease: • Angelman Syndrome (aberrant DNA methylation) • Beckwith-Weideman syndrome (aberrant DNA methylation) Suboptimal culture conditions may be a causative factor for predisposing offspring to these syndromes M Bi M un-M Cause for biallelic expression was due to aberrant DNA methylation Animals produced by Somatic Cell Nuclear Transfer in particular, can have very profound phenotypes… Pace et al., Biol Rep 67, 2002 In vitro fertilization Cezar et al., Biol Rep 68, 2003 SCNT(cloning) control Tamashiro et al., Nat Med 8(3), 2002 The general consensus is that the cause of these phenotypes are EPIGENETIC in nature • Epigenetics is a mechanism that ensures heritable characteristics of cells and functional differences between cell types • Epigenetic mechanisms alter chromatin (DNA and proteins) in ways that change the availability of genes to transcription factors. Key components include: – Addition of methyl group to CpG dinculeotides* – Association of Polycomb and other DNA binding proteins that modify histones. DNA methylation patterns • Known to be established during development and subsequently maintained by DNA methyltransferases • Traditionally thought that once established, the methylation patterns were reliably maintained for the life of the organism and irreversible. – View is evolving a bit these days. Are Imprinted Genes Differentially Expressed in Response to Maternal Under-Nutrition? What is an imprinted gene? • Genes that are mono-allelically expressed -Either maternal allele OR paternal allele is expressed • Paternal genes are thought to extract maternal resources for the benefit of the offspring – Growth promoters • Maternal genes are thought to allocate resources ‘equitably’ between offspring and mother. – Growth suppressors • Thus, imprinted genes have growth related functions with antagonistic properties. • Imprinted gene expression is epigenetically regulated. Facts about Imprinted Genes • ~70-200 exist in mammals • Found in clusters (referred to as imprinted domains) at ~ 15 different chromosomal sites • Imprinted domains are coordinately regulated by epigenetic mechanisms: – DMD, or Differentially Methylated Domain H19 and Igf2: Examples of imprinted genes and coordinate regulation of expression through a differentially methylated domain. Experimental design: C57B6 female mice Mating pcd5 2 wks prior to mating C57B6 females fed a low protein Diet. pcd10 pcd19 Pups delivered by C-section; Organs harvested for mRNA isolated to measure expression levels of imprinted genes in each individual organs (n=10). Controls fed normal chow diet throughout. LPD diet Normal chow diet Imprinted Genes: Igf2 Igf2r H19 Ata3 P57kip2 Peg1/Mest Peg3 Allele Expressed Function (based on observed phenotypes found in mutations). Paternal Maternal Maternal Paternal Maternal Paternal Paternal Growth Growth retardation ?? Amino acid transport (Placenta) Reduced survival; growth retardation Growth Growth Imprinted Genes: Liver H19 18 16 14 12 10 8 6 4 2 0 Con Brain Heart Kidney ND ND ND Exp 3 0.8 3.5 3 Igf2 2.5 0.7 0.6 0.5 2 0.4 1.5 1.5 0.3 1 1 0.2 0.5 0.1 0 0 Con 0.5 0 Con Exp ND 2 2.5 Exp Con Exp 3.5 3 3 2.5 2.5 2 Igf2r 2 ND 1.5 1 0.5 ND 1.5 1 0.5 0 0 Con Con Exp Exp 2 1.85 1.8 1.5 1.75 Peg3 ND 1.7 1.6 0.5 1.55 1.5 0 1.45 1.4 Con Ata3 ND 1 1.65 ND Exp ND Con ND 3 Exp 2 1.8 1.6 1.4 1.2 1 0.8 0.6 0.4 0.2 0 Con Exp 2.5 2 P57kip2 ND 1.5 ND 1 0.5 0 Con Exp ND Is Altered Expression of H19 Gene Expression Associated with a Change in Methylation of the DMD? Igf2 H19 26 CpG dinucleotides? Summary • Exposure to a maternal LPD during preimplantation period appears to alter imprinted gene expression in organs of near born pups. • Expression appears to be tissue specific • No detected differences in methylation patterns of DMD • Encouraged by the preliminary data indicting imprinted gene expression, known to be regulated epigenetically, could be altered by maternal nutrition. Submitted a Pilot and Feasibility Grant to the CNRU. CNRU P & F Proposal Weaning* 6 wks. 9 wks.* 12 weeks Normal Chow Low Protein Pups weaned to Hi fat Low Protein Pups weaned to Low protein Hi Fat Pups weaned to Hi Fat Can we link later in life phenotypic characteristics such as weight, BP, glucose tolerance, diabetes,etc. with differentially expressed genes and methylation patterns of associated CpG islands and/or promoters? Acknowledgements Ms. Heather Kirk Dr. Barry Robert (Kidney) Ms. Regina Staten Stem Cell Biology Lab: Dr. Jeff Gimble Dr. Randy Mynatt Dr. Basia Kozak Dr. Beth Floyd Dr. Rob Koza: Bisulphite Sequencing Low Density Arrays Microarrays !Dr. Eric Ravussin!